Difference between revisions of "Amarel"
| (190 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<div class="title">Using Amarel</div> | <div class="title">Using Amarel</div> | ||
==Getting Started== | |||
This wiki page is a brief "Getting Started" introduction to running the ROMS ocean model and analyzing model output on the Rutgers University Office of Advanced Research Computing (OARC) cluster computer ''Amarel''. | |||
===Amarel help === | |||
There is an OARC supported user guide at https://sites.google.com/view/cluster-user-guide to which you can refer for a lot of detailed guidance. | |||
===Working with UNIX=== | |||
To work effectively on ''Amarel'' you need at least a beginner level of competency with the UNIX operating system. | |||
If you are a total newcomer to UNIX computers you might find it useful to work through a structured tutorial; for example [https://ryanstutorials.net/linuxtutorial/ Ryan's Linux Tutorial]. | |||
{| class="wikitable" | |||
|+ Basic UNIX commands | |||
|- | |||
! Command !! What it does !! Example | |||
|- | |||
| ls || list contents of a directory </br> files ending in .nc </br> long format list and hidden files || ls </br> ls *.nc </br> ls -al | |||
|- | |||
| cd || change directory || cd /projects/dmcs_1/courses/cod | |||
|- | |||
| pwd || print working directory to see where you are || pwd | |||
|- | |||
| cp || copy a file || cp source_file target_file | |||
|- | |||
| mv || rename a file </br> move file to new directory || mv file_name new_name </br> mv file_name dir1/dir2/new_name | |||
|- | |||
| rm || delete (remove) a file || rm filename | |||
|- | |||
| more || type contents of a file to screen one page at a time || more .bashrc | |||
|- | |||
| less || like ''more'' only with more features (unix geeks are so funny) </br>Read the man page for help || less .bashrc | |||
|- | |||
| man || get the manual page (help) for a command || man less | |||
|- | |||
| . || the directory you are in || cp /projects/dmcs_1/courses/cod/job.sh '''.''' | |||
|- | |||
| .. || the directory above the one you are in || cd '''..''' | |||
|- | |||
| grep || find a string in a file </br> ''get regular expression and print (!)'' || grep COD_1DMIX *.h | |||
|} | |||
Your default UNIX ''shell'' (the "flavor" of your UNIX environment) is '''bash''', so for more advanced help it is often good to include "bash" as one of your search terms in a google search for help. | |||
===Editing files=== | |||
One of the things that stumps newcomers to UNIX is making simple edits to files. Ryan's Linux Tutorial goes over the basics of the '''vi''' editor [https://ryanstutorials.net/linuxtutorial/vi.php here], but it can be a beast to get comfortable with. | |||
A more intuitive editor for new users is '''nano''', the basics of which you can read about [https://www.howtogeek.com/howto/42980/the-beginners-guide-to-nano-the-linux-command-line-text-editor/ here]. | |||
There is also a way to edit files in the web browser based '''OnDemand''' service that is described later in this guide. | |||
==Rutgers VPN== | ==Rutgers VPN== | ||
If you are not on campus, you will | If you are not on campus, you will need to connect to the Rutgers VPN. To use Rutgers VPN, you must first enroll in Duo 2 Factor Authentication (2FA) (NetID+). Most users have probably already done this. If not, navigate to https://netid.rutgers.edu/setupTwoFactorAuthentication.htm and follow the instructions. | ||
Once you are enrolled in NetID+ you will need to activate the VPN service, if you have not already. Navigate to https://soc.rutgers.edu/vpn/ and click the gray button titled '''Service Activation'''. If you are not already logged in you may be asked to login and/or approve | Once you are enrolled in NetID+ you will need to activate the VPN service, if you have not already. Navigate to https://soc.rutgers.edu/vpn/ and click the gray button titled '''Service Activation'''. If you are not already logged in you may be asked to login and/or approve your login with the Duo Mobile app. To activate the VPN service, click the checkbox next to '''Remote Access VPN, Cisco AnyConnect Access for Rutgers''' and click the '''Activate Services''' button. | ||
You are now ready to download the Cisco VPN client and connect to the Rutgers VPN. Complete instructions can be found [https://soc.rutgers.edu/vpn/ here] by clicking the red button titled '''General VPN Instructions''' to download the PDF. The instructions are geared towards Windows so if you are using a Mac you might find [https://soc.rutgers.edu/vpn/apple/ this page] more helpful. In most cases, regardless of your operating system, pointing your browser to https://vpn.rutgers.edu/ and logging in with your NetID will lead you to downloading the correct VPN client. | You are now ready to download the Cisco VPN client and connect to the Rutgers VPN. Complete instructions can be found [https://soc.rutgers.edu/vpn/ here] by clicking the red button titled '''General VPN Instructions''' to download the PDF. The instructions are geared towards Windows so if you are using a Mac you might find [https://soc.rutgers.edu/vpn/apple/ this page] more helpful. In most cases, regardless of your operating system, pointing your browser to https://vpn.rutgers.edu/ and logging in with your NetID will lead you to downloading the correct VPN client. | ||
| Line 11: | Line 61: | ||
[[Image:AnyConnect_Connect.png]] | [[Image:AnyConnect_Connect.png]] | ||
In the next window, | In the next window, shown below, '''Username''' will be your NetID. | ||
In the second line '''Password''' is the password for your NetID. | |||
The third line is the '''Duo Action''' to complete the 2FA, which can be accomplished in one of 4 possible ways: | |||
# Enter a 6 digit Duo Passcode. These are generated either by a Hard Token, by showing the passcode in the Duo Mobile App, or from a previous “SMS” request. Simply type in the 6 numbers and hit OK. | |||
# Type the word “push”. This will send a push notification to the primary device you have enrolled with Duo through NetID+ with the option to Accept or Deny. | |||
# Type the word “phone”. You will receive a phone call to the primary device you have enrolled with Duo through NetID+ with touch tone options to Accept or Deny. | |||
# Type the word “sms”. You will receive a text message to the primary device you have enrolled with Duo through NetID+ containing passcodes you can use to login. | |||
[[Image:AnyConnect_Credentials.png]] | [[Image:AnyConnect_Credentials.png]] | ||
| Line 24: | Line 77: | ||
==Connecting to Amarel with SSH== | ==Connecting to Amarel with SSH== | ||
In order to connect to | In order to connect to ''Amarel'' and compile and run ROMS you will need an SSH/terminal client. If you already use an SSH/terminal application you are comfortable with then stick with that and adapt the instructions below accordingly. For these instructions to work you need to either be on campus or connected to the Rutgers VPN. | ||
===Mac OS=== | ===Mac OS=== | ||
If you are on a Mac, open Terminal (found in the Applications -> Utilities folder) or | If you are on a Mac, open the '''Terminal''' app (found in the Applications -> Utilities folder) or '''iTerm''' app and type: | ||
:<div class="box">ssh fakeuser@amarel.rutgers.edu</div> | |||
replacing <span class="blue">fakeuser</span> with your NetID. When it asks if you want to continue connecting enter '''yes''', then enter your NetID password. | |||
Having to type the above command and netid password every time you login is moderately tedious, but this username/password authentication can become annoying and time consuming when repeatedly using the '''scp''' command to copy files to ''Amarel''. Passwordless access and file transfer can be enabled using '''SSH keys''' by following the instructions below, or for more detail see the [https://sites.google.com/view/cluster-user-guide#h.jgwrkm9e9rwg OARC User Guide on this topic]. | |||
# If you are still connected to ''Amarel'', disconnect by entering '''exit''' | |||
# At the prompt for your local machine, enter '''<span style="font-family: monospace;">nano ~/.ssh/config</span>''' and paste the block below at the end of the file:<div class="box">Host amarel<br />Hostname amarel.rutgers.edu<br />HostKeyAlias amarel<br />User NetID</div> replacing <span class="blue">NetID</span> with your own NetID. Save and exit (Ctrl+o Ctrl+x). This allows you to type '''<span style="font-family: monospace;">ssh amarel</span>''' to connect instead of '''<span style="font-family: monospace;">ssh fakeruser@amarel.rutgers.edu</span>''' | |||
# Enter '''<span style="font-family: monospace;">ls ~/.ssh</span>'''. If there is a file listed called <span class="blue">id_rsa.pub</span> skip to 5 | |||
# Enter '''<span style="font-family: monospace;">ssh-keygen -t rsa</span>''', then hit return to accept the default location, enter a passphase twice (or we recommend you just leave it blank twice for no passphrase) | |||
# Copy the public portion of the key to ''Amarel'' by entering '''<span style="font-family: monospace;">scp ~/.ssh/id_rsa.pub amarel:.</span>''' | |||
# SSH to ''Amarel'' ('''<span style="font-family: monospace;">ssh amarel</span>''') and check if you already have a .ssh folder:<div class="box">[rjdave@amarel1 ~]$ ls -l .ssh<br />total 33<br />-rw------- 1 rjdave rjdave 1069 Sep 8 2017 authorized_keys<br />-rw-r--r-- 1 rjdave rjdave 434 Jun 4 2021 config<br />-rw-r--r-- 1 rjdave rjdave 5992 Dec 2 13:47 known_hosts</div> You probably won't have all the files above but if you instead get the message '''<span style="font-family: monospace;"><div class="box">ls: cannot access .ssh: No such file or directory</span>'''</div>then enter the following command:</br>'''<span style="font-family: monospace;">mkdir .ssh && chmod 700 .ssh</span>'''. </br>These two commands (combined on one line) will create the .ssh folder and set the required permissions. Setting private user-only permissions ('''chmod 700 ...''') is important, because the system will not allow you to have your keys viewable by others users. | |||
# Once you have a .ssh folder with the proper permissions, enter the following commands</br> '''<span style="font-family: monospace;">cat ~/id_rsa.pub >> ~/.ssh/authorized_keys && chmod 640 ~/.ssh/authorized_keys</span>'''</br>then exit back to your local machine and execute '''<span style="font-family: monospace;">ssh amarel</span>''' again. It should no longer ask for your password. | |||
===Windows=== | ===Windows=== | ||
| Line 44: | Line 102: | ||
==Setting up your .bashrc== | ==Setting up your .bashrc== | ||
The | The default software setup by OARC on ''Amarel'' does not include everything needed to compile and run ROMS. Ocean Modeling Group computing specialist David Robertson has configured, and maintains and updates, a repository of what you need for ROMS and other models that will be automatically loaded for every login session once you add some lines to the login script (.bashrc, .cshrc, etc.) in your home directory. Log in to ''Amarel'' with '''Terminal''', '''iTerm2''' or '''MobaXterm''' and edit your login script as described below. | ||
Unless otherwise requested your default shell will be bash and the following lines shown below in <span class="red">red</span> and <span class="forestGreen">green</span> should be added (with nano or your preferred editor) near the top of your .bashrc after the import of the global definitions as shown: | |||
:<div class="box"># Source global definitions<br />if [ -f /etc/bashrc ]; then<br /> . /etc/bashrc<br />fi<br /><br /><span class="red">ulimit -s unlimited<br />export MODULEPATH=/projects/dmcs_1/sw/modulefiles/Core:${MODULEPATH}<br />export SQUEUE_FORMAT="%.18i %.9P %.8j %.8u %.2t %.10M %.10l %.4C %.6D %R"</span><br /><br /><span class="forestGreen"># Automatically load the roms module<br />module load roms</span></div> | |||
The line with <span class="red">ulimit</span> allows you to take full advantage of computing resources (may be obsolete now but it won't hurt anything). | |||
The first <span class="red">export</span> line will tell '''Lmod''' where to find the modules that will load the custom software. | |||
The second <span class="red">export</span> line makes the '''squeue''' command (explained later) more useful. | |||
The lines in <span class="forestGreen">green</span> are optional and will load the '''roms''' module automatically every time you login to ''Amarel''. They must be '''after''' the line that sets the '''MODULEPATH'''. | |||
Once you have added these lines, log out and back in for them to take effect. | |||
{{note}} '''Note:''' If you see the error ... | |||
:<div class="box"><span class="red">Lmod has detected the following error:</span> The following module(s) are unknown: "roms"<br /><br />Please check the spelling or version number. Also try "module spider ..."<br />It is also possible your cache file is out-of-date; it may help to try:<br /> $ module --ignore_cache load "roms"<br /><br />Also make sure that all modulefiles written in TCL start with the string #%Module</div> | |||
... then you may need to logout and login again so that .bashrc sets MODULEPATH as instructed above. If the error persists, you may not have been properly added to the '''dmcs_1''' Linux group. To check if you are part of '''dmcs_1''' type the command '''<span style="font-family: monospace;">id</span>''' and if '''dmcs_1''' is not listed, let us know and we will get you added to the group. | |||
==Accessing the ROMS Source Code== | |||
===Register as a ROMS user=== | |||
If you are not already a ROMS user, you will need to fill out the registration form found [https://www.myroms.org/register here] and wait for approval. Once approved, you will be able to checkout the ROMS source code. It is highly recommended that you use the GitHub myroms repository to obtain the ROMS source code. The ''Subversion (SVN)'' repository is being phased out and will no longer be available after '''December 31, 2024'''. If you manage your own software projects using GitHub, the concepts for software version control may already be familiar to you. Detailed directions can be found on the WikiROMS [[Git#Downloading_ROMS|Git]] page. | |||
===Obtaining the ROMS source code=== | |||
There is nothing special about obtaining the ROMS source code on ''Amarel''. The git commands are the same as on any UNIX host. | |||
Navigate to the directory where you want your ROMS source code to reside and execute this git clone command: | |||
:<div class="box">git clone https://github.com/myroms/roms.git <my_src_dir></div> | |||
replacing <my_src_dir> with what you want the source code directory to be called. You can also omit <my_src_dir> and git will automatically name the folder '''roms'''. | |||
It can take a couple of minutes to download all of ROMS, but you only do this once. | |||
Many users simply run this command from their home directory and use <span class="forestGreen">'''myroms'''</span> for <my_src_dir>, but the choice is yours depending on how you want to manage your ''Amarel'' space and how many ROMS projects you think you might end up working on. | |||
==Loading and Unloading Modules== | |||
Like many clusters, ''Amarel'' uses environment modules to load and unload software and configure the environment. Some commands you will find useful are: | |||
:<div class="box">'''module help''' Display help message<br />'''module help <m1'''> Show help information for module <m1><br />'''module available''' Show all modules currently available<br />'''module whatis <m1>''' Show brief information about module <m1><br />'''module spider''' List all possible modules even if not currently available<br />'''module list''' List currently loaded modules<br />'''module load''' Load the specified module(s)<br />'''module unload''' Unload the specified module(s)<br />'''module swap <m1> <m2>''' Unload <m1> and then load <m2> (for switching versions of the same software)<br />'''module purge''' Unload all loaded modules</div> | |||
===Loading the ROMS Module=== | |||
If you '''did not''' add the optional lines from the [[#Setting_up_your_.bashrc|Setting up your .bashrc]] section, you can easily set up your environment to compile and run ROMS by manually loading the '''roms''' module. | |||
:<div class="box">module load roms<br />module list<br />Currently Loaded Modules:<br /> 1) intel/17.0.4 2) mvapich2/2.2 3) mct-roms/2.6.0 4) netcdf/4.6.2<br /> 5) esmf/8.0.0_nc4 6) parpack-roms/2.1 7) hdf5/1.10.4 8) roms/intel_nc4</div> | |||
Notice that loading the '''roms''' module will actually load 7 other modules. | |||
This will setup your environment to use the Intel compiler so remember to set '''FORT''' to '''ifort''' in your build script. | |||
{{note}} '''Note:''' If you see the error ... | |||
:<div class="box"><span class="red">Lmod has detected the following error:</span> The following module(s) are unknown: "roms"<br /><br />Please check the spelling or version number. Also try "module spider ..."<br />It is also possible your cache file is out-of-date; it may help to try:<br /> $ module --ignore_cache load "roms"<br /><br />Also make sure that all modulefiles written in TCL start with the string #%Module</div> | |||
... you may not have been properly added to the '''dmcs_1''' Linux group. To check if you are part of '''dmcs_1''' type the command '''<span style="font-family: monospace;">id</span>''' and if '''dmcs_1''' is not listed, let us know and we will get you added to the group. | |||
==Your project/coursework working directory space== | |||
We recommend you create a subdirectory of your home directory in which to work, so you can keep each ROMS project separate and organized. | |||
You might call this '''working directory''' <span class="forestGreen">CoastalDynamics</span>, or <span class="forestGreen">Projects</span>, or simply <span class="forestGreen">cod</span> (for coastal ocean dynamics). You will be typing this a lot, so keep it simple. | |||
To make a new directory called <span class="forestGreen">Projects</span>, enter this command | |||
:<div class="box">mkdir Projects </div> | |||
Change to this working directory with the command | |||
:<div class="box">cd Projects </div> | |||
In instructions that follow, when we refer to your '''working directory''' we mean this workspace. | |||
==Configuring and Compiling ROMS== | |||
You will compile and run ROMS in your project/coursework '''working directory''', so change to that directory first. | |||
:<div class="box">cd <span class="forestGreen">Projects</span> </div> | |||
Running the build script <span class="blue">build_roms.sh</span> is the recommended method for compiling ROMS on ''Amarel''. For any given project you will likely only need to change <span class="blue">build_roms.sh</span> to reset <span class="sandyBrown">ROMS_APPLICATION</span> and <span class="sandyBrown">MY_ROMS_SRC</span> and make sure <span class="sandyBrown">FORT</span> is set to '''ifort'''. | |||
{{warning}} <span class="red">It is important to make sure '''USE_MY_LIBS''' is set to ''no'' or your compilation will fail.</span> | |||
A template <span class="blue">build_roms.sh</span> script is in subdirectory '''ROMS/Bin''' of '''<my_src_dir>''' that you created when you downloaded the code. For example, if you created <span class="forestGreen">myroms</span> in your home directory the copy command would be | |||
:<div class="box">cp ~/myroms/ROMS/Bin/build_roms.sh . </div> | |||
Now open <span class="blue">build_roms.sh</span> with an editor (''e.g.'' '''nano''') and modify the line <span class="sandyBrown">MY_ROMS_SRC</span> to point to your choice for <my_src_dir>. So, you will have something like ... | |||
:<div class="box"><span class="sandyBrown">export MY_ROMS_SRC=</span>{$HOME}/<span class="forestGreen">myroms</span> </div> | |||
if the name you chose for <my_src_dir> was <span class="forestGreen">myroms</span>. | |||
If this is your first time working with ROMS, a good starting place is to compile the default UPWELLING test case that is indicated in build_roms.sh with the default setting <span class="sandyBrown">export ROMS_APPLICATION</span>=UPWELLING. | |||
To compile the UPWELLING example, you need to copy the configuration file <span class="blue">upwelling.h</span> to your working directory. This is in subdirectory '''ROMS/Include''' of your source code. For example, the command might be: | |||
:<div class="box">cp ~/myroms/ROMS/Include/upwelling.h .</div> | |||
Once your build script is configured and you have upwelling.h in your working directory you can compile ROMS by typing: | |||
:<div class="box">./build_roms.sh -j 4</div> | |||
where the number after -j indicates the number of compute cores to use in parallel to execute the compilation. The greater the number, the faster it goes. | |||
However, the login node you will be compiling on is shared for the entire ''Amarel'' system. If you use a number larger than '''4''', or omit it altogether (which says use ''all'' cores on the login node) your build might be terminated by an administrator. Be a considerate user and keep the number low. | |||
If compilation is processing successfully you will see a lot of information typed to your screen as each component of the source code is compiled. It will take a minute or so to complete. | |||
If compilation ends quickly with errors like this: | |||
:<div class="box">$ ./build_roms.sh -j 4 </br>rm -f -r core *.ipo /home/jwilkin/cod/Projects/Build_roms...</br>which: <span class="red">no mpif90</span> in (/usr/lpp/mmfs/bin:/usr/local/bin:/us...</br>cp -f /opt/intelsoft/serial/netcdf3/include/netcdf.mod /h...</br>cp: cannot stat ‘/opt/intelsofo... No such file or directory</br>make: *** No rule to make target netcdf.mod needed by ... Stop. </div> | |||
... this means you have not run the <span class="blue">module load roms</span> command. Go back to the notes on [[Amarel#Loading_the_ROMS_Module]]. | |||
If compilation was successful, there will be a file named '''romsM''' that is the model executable. You can check for the file with this command: | |||
:<div class="box">ls -l *</div> | |||
You should see: | |||
:<div class="box"><span class="blue">Build_roms</span> <span class="forestGreen">build_roms.sh romsM</span> upwelling.h </div> | |||
<span class="blue">Build_roms</span> is a folder that build_roms.sh created to hold all the temporary files that the FORTRAN compilation process makes to combine into the romsM executable file. | |||
==Running on the Amarel Compute Nodes== | |||
When you '''ssh''' to ''Amarel'' you are connected to one of the login nodes. These nodes are to be used for file editing, transfering output and input files to/from your local computer, and modest code compiling tasks and analysis, but not for compute intensive tasks. Here we explain how to connect to the compute nodes for these larger tasks. | |||
Note: Consult the Amarel status page (https://oarc.rutgers.edu/amarel-system-status/) before scheduling a job. Amarel is down for maintenance monthly. | |||
===What ''partitions'' can you run on?=== | |||
Access to Amarel resources is controlled by ''partitions''. You can find out the partitions you have permission to access with the command | |||
:<div class="box">sinfo -s</div> | |||
This will show the partitions that everyone has access to. See the cluster guide [https://sites.google.com/view/cluster-user-guide#h.oeejy9yf80e4 entry on partitions] for an explanation of these. | |||
If you belong to a research group that own a partition you should also see that listed, for example '''p_omg_1''' for the Ocean Modeling Group. This is the partition you want to use most of the time as your job will not get pre-empted by other owner users. | |||
===Using the SLURM batch job scheduler=== | |||
''Amarel'' uses SLURM workload manager to schedule compute intensive tasks. The user configures a SLURM job script for each model run and submits this script with the '''sbatch''' command. The job script declares the resources required, such as number of CPUs for parallel jobs, maximum memory required, etc. | |||
We have configured a simple template job script (for the ROMS UPWELLING example) that you can copy from '''/projects/dmcs_1/courses/cod/job.sh''' into your working directory. | |||
The UNIX command to copy a file is '''cp source_file target_file'''. So, you would enter | |||
:<div class="box">cp /projects/dmcs_1/courses/cod/job.sh .</div> | |||
The syntax here uses a ''dot'' ('''.''') for the target_file, which causes the source_file (job.sh) to be copied to the present working directory without changing its name. | |||
When typing this, or any, UNIX command, you can start typing just the beginning few characters of the path then press TAB to autocomplete the filename. If the completion is ambiguous, you can continue typing more characters and press TAB again. Pressing TAB twice in a row will show you the set of possible completions. This trick can save you a lot of tedious (mis)typing. | |||
The contents of '''job.sh''' are shown below: | |||
:<div class="box">#!/bin/bash<br >#SBATCH --partition=p_omg_1 # Partition. If you are not a member of OMG enter your own partition<br />#SBATCH --job-name=upwelling # Assign a short name to your job<br/>#SBATCH --nodes=1 # Number of nodes you require<br />#SBATCH --ntasks=4 # Total number of tasks you'll launch<br />#SBATCH --ntasks-per-node=4 # Number of tasks you'll launch on each node<br />#SBATCH --cpus-per-task=1 # Cores per task (>1 if multithread tasks)<br />#SBATCH --mem=6400 # Real memory (RAM) required (MB) per node<br />#SBATCH --time=00-00:05 # Total run time limit (DD-HH:MM)<br />#SBATCH --output=out.%N.%j # STDOUT output file<br />#SBATCH --error=err.%N.%j # STDERR output file (optional but recommended)<br />#SBATCH --export=ALL # Export you current env to the job env<br /><br />## It is important to have --mpi=pmi2 here or ROMS will not run<br />srun --mpi=pmi2 ./romsM roms_upwelling.in</div> | |||
Note: The maximum run time is 14-00:00 (14 days). It is tempting to set this to the maximum in order to avoid a job ending too early. (''i.e.'' Slurm stopping a ROMS run before it is finished) However, if there is maintenance scheduled during that time period the job will not start until maintenance is complete. So a runtime as close as possible to the actual run time is recommended. | |||
====Requesting a specific class of compute node==== | |||
For large jobs it makes sense to use all available cores on a node, and for low priority jobs to use older less fast nodes. You can do this by requesting a specific node number, but it is better to request a particular class of nodes so you don't unwittingly pick a node already in use. The classes available from oldest to newest nodes are: | |||
# skylake (32 cores) (Phase I nodes) (nodes hal0116 through hal0120) Note: These will be reclaimed for general use in Feb 2024 | |||
# cascadelake (40 cores) (Phase II nodes) (nodes hal0159 through hal0163) | |||
# icelake (64 cores) (Phase V.B nodes) (nodes hal0299 -- hal0302 and hal0346 -- hal0351) | |||
Make the node class request in the SLURM script and choose the maximum number of cores to match, ''e.g.'' | |||
:<div class="box">#SBATCH --constraint=cascadelake # Run on Phase II nodes (40 cores) <br>#SBATCH --ntasks=40 # Total number of tasks to launch (like -np to mpirun) <br>#SBATCH --ntasks-per-node=40 # Number of tasks to launch on each node | |||
</div> | |||
If you don't specifically request a class of nodes the SLURM scheduler will make its own choice on the most appropriate resources to use given your request. We don't know the basis of that choice. | |||
If you request a class of node that is fully assigned to other users in your partition, your job will wait for the others users jobs to complete. So, coordinate with your colleagues if resource use is at a premium. You can use the '''squeue -p partition_name''' command to see what resources are being used by other users, ''e.g.'' | |||
:<div class="box">$ squeue -p p_omg_1 <br> JOBID PARTITION NAME USER ST TIME TIME_LIMIT CPUS NODES NODELIST(REASON) <br> 26975473 p_omg_1 fwd levinj R 17:15:42 14-00:00:00 80 2 hal[0159,0162] | |||
</div> | |||
===Running the ROMS Upwelling Example=== | |||
You may have noticed that the srun command above includes a file named <span class="blue">roms_upwelling.in</span>. You will need to copy this file and <span class="blue">varinfo.yaml</span> from the ROMS/External directory of your <my_src_dir> ROMS source code to the directory where you compiled ROMS. | |||
If, for example, you chose '''myroms''' as the name of your <my_src_dir> when you downloaded the code, you would enter | |||
:<div class="box">cp ~/myroms/ROMS/External/roms_upwelling.in .</div> | |||
The syntax here uses ''tilde'' (~) as shorthand for your home directory, | |||
After you copy the files, you will need to make a couple of small edits (using ''e.g.'' '''nano''') to <span class="blue">roms_upwelling.in</span> to get this to work. | |||
# Change the line with VARNAME to read <span class="sandyBrown">VARNAME = varinfo.yaml</span> (''i.e.'' delete the ROMS/External part) | |||
# Set <span class="sandyBrown">NtileI == 2</span> and <span class="sandyBrown">NtileJ == 2</span>. | |||
The product of NtileI and NtileJ, ''i.e.'' 2 x 2 = 4, is the number of cores the model will run on in parallel. This number must match the number in the SLURM job script options '''--ntasks''' and, in this case, '''--ntasks-per-node''' | |||
Now you can submit your job to '''sbatch''' with the command '''<span style="font-family: monospace;">sbatch job.sh</span>''' | |||
===ROMS configurations for ''Coastal Ocean Dynamics'' class=== | |||
The files that will configure the examples we will study in class will be located in <span class="blue">/projects/dmcs_1/courses/cod</span>. | |||
You will find instructions for configuring and running the examples at a separate Wiki page [[COD_Class_Examples]] | |||
===Checking Job Status with squeue=== | |||
Detailed documentation for monitoring your SLURM jobs can be found [https://sites.google.com/view/cluster-user-guide#h.4bsndqufii8p here]. The easiest way to check whether your job is running is with the command '''<span style="font-family: monospace;">squeue -p p_omg_1</span>''' (or your own partition name if it is not p_omg_1). | |||
You should see output something like this: | |||
:<div class="box"> JOBID PARTITION NAME USER ST TIME TIME_LIMIT CPUS NODES NODELIST(REASON)<br /> 17197219 dmcs_1 watl_psas rjdave R 3-04:07:00 3-20:00:00 4 1 hal0035<br /> 17146609 dmcs_1 upwelling rjdave R 00:01:10 00:05:00 4 1 hal0035</div> | |||
You can see that the job is running (the ‘R’ under ‘ST’), has been running for 1 minute, 10 seconds, and is running on node 35. | |||
===Checking Progress with tail=== | |||
You can check ROMS progress by using the tail command on the '''output''' file. For the upwelling job above the file would be called <span class="blue">out.hal0035.17146609</span> so the command '''<span style="font-family: monospace;">tail out.hal0035.17146609</span>''' will show you the most recent 10 lines written to the output log and will most likely tell you what time-step the model is on. Using the -f option ('''<span style="font-family: monospace;">tail -f out.hal0035.17146609</span>''') will output appended data as the file grows. Ctrl-C will escape the display. | |||
===Did my UPWELLING job run?=== | |||
If the UPWELLING test case was successful there should be an output log file named something like <span class="blue">out.hal0035.17146609</span> that shows no errors, and there should be 4 output NetCDF files ... | |||
:<div class="box">ls -l *.nc </br>roms_avg.nc </br>roms_dia.nc </br>roms_his.nc </br>roms_rst.nc </div> | |||
===Cancelling a Job=== | |||
To safest way cancel a queued or running job is to use its jobid. A job can be canceled by name but that is not recommended. To cancel the upwelling job in the example above you would issue the command '''<span style="font-family: monospace;">scancel 17146609</span>'''. If the job has not yet started, it will be removed from the queue. If it is running, all child processes will be killed and the job will be removed from the queue. You are only able cancel jobs that you own. | |||
===Running an interactive session on a compute node=== | |||
It is possible to conduct your work interactively on one of the compute nodes (instead of the login node). For most work we will be doing in class this is not necessary, but if for some reason you have a job that needs many processors or a large amount of memory, and you want to run it interactively - say, to simply check that everything is in order for it to start correctly - there are instructions in the Cluster User Guide [https://sites.google.com/view/cluster-user-guide#h.26x9sbburvsg here]. | |||
For compute intensive interactive work, such as model analysis using Python or MATLAB, we recommend using the OnDemand interface to launch an interactive session on a set of compute nodes. | |||
==Using the OnDemand desktop environment== | |||
The '''OnDemand''' service allows you to connect to ''Amarel'' using a graphical desktop environment in a web browser. One of the most useful features of '''OnDemand''' is that it allows you to analyze model output by Python or Matlab on the cluster compute nodes. | |||
You need to be running a Rutgers VPN to use OnDemand. | |||
Navigate your browser to https://ondemand.hpc.rutgers.edu/pun/sys/dashboard and login with your NetID and password. At the top of the page click '''Interactive Apps'''. | |||
===Python Jupyter notebook:=== | |||
# Select '''Personal Jupyter''' from the list on the left | |||
# Make your selection for '''number of hours''' your session can remain active (''e.g.'' <span class="blue">3</span>) hours | |||
# Set the '''number of cores''' to run on (I suggest <span class="blue">1</span> for now) | |||
# Set '''memory''' (<span class="blue">8</span> GB is an OK choice) | |||
# For ''Partition'' enter ''e.g.'' <span class="blue">p_omg_1</span> | |||
# Leave ''Reservation'' blank | |||
# For ''Slurm feature'' enter <span class="blue">cascadelake</span> so your session starts on our older nodes and does not interfere take the larger, faster cores being used by OMG | |||
# For ''conda path'' enter <span class="blue">/projects/dmcs_1/miniconda3</span> | |||
# For ''conda environment'' enter <span class="blue">/projects/dmcs_1/sw/packages/xroms/py38</span>. This '''conda''' environment built for DMCS_1 makes available many of the Python modules you are used to using. | |||
These settings are remembered the next time you access OnDemand. | |||
Click '''Launch''' and wait. This can take a couple minutes. | |||
You should see sub-window of the web page like this </br> [[Image:JupyterStarting.png|600px]] </br></br>When the session is ready it changes to this: </br> [[Image:JupyterRunning.png|600px]] | |||
Start your notebook by clicking the '''Connect to Jupyter, Anaconda version 5.1.0''' button: (If this does not appear, check that you correctly entered the '''conda path''' and '''conda environment''' at step 3 above.) </br>[[Image:JupyterConnect.png|400px]] | |||
This should place you in a file browse window from which you can navigate to one of your existing Jupyter notebooks, or, to start a new Untitled notebook, select ''Python 3 (ipykernel)'' from the ''New'' pull-down menu on the right side of the page [[Image:NewPythonNotebook.png]] | |||
Once your notebook is running, you can import the xroms package as you would any other toolbox, by entering '''<span style="font-family: monospace;">import xroms</span>''' then hold down the shift key and hit the return/enter key. You will see an asterisk ['''*'''] in the square brackets. | |||
At present the '''<span style="font-family: monospace;">import</span>''' steps are quite slow ... 30 seconds ... so be patient and don't re-run those cells in your notebook if you don't have to. | |||
===Matlab:=== | |||
# Select MATLAB from the list on the left | |||
# As for Step 2 above for Python, choose time, cores, and memory | |||
# Select MATLAB version, and enter <span class="blue">p_omg_1</span> (or your own group name) in partition. | |||
# Click '''Launch''' and wait. This can take a couple minutes. | |||
# Once the '''Launch noVNC in New Tab''' button appears, click it and a MatLab GUI will open. | |||
You are running in a graphical desktop environment, so by default Matlab will look in Documents/MATLAB/startup.m for your personal Matlab configuration file. | |||
Popular MATLAB utilities for working with ROMS output are in <span class="blue">/projects/dmcs_1/sw/packages/matlab-tools</span>. These include the myroms.org utilities in subdirectory <span class="blue">roms_matlab</span>, and John Wilkin's toolbox in <span class="blue">roms_wilkin</span>. | |||
===Reconnecting to an interactive session=== | |||
A particularly nifty feature of '''OnDemand''' is that your interactive session is independent of the browser window instance. | |||
Say you are working on your office computer using a Jupyter notebook via '''OnDemand''' but haven't finished all you want to do by the end of the day. If you leave that session running, you can go home and connect once again to '''OnDemand''' and you will see your session in the list of Interactive Sessions. Launch that session and you will be placed right back where you were. '''OnDemand''' is simply a web interface to code running on ''Amarel'', so you can connect from any browser. | |||
Of course, this only works if you selected a '''number of hours''' in Step 2 above that will allow your session to remain open. | |||
As long as the session is active the ''Amarel'' resources (cores and memory) are reserved for you and no one else. So, be respectful of other users and close out an interactive session once you are done. | |||
Latest revision as of 16:00, 6 February 2024
Getting Started
This wiki page is a brief "Getting Started" introduction to running the ROMS ocean model and analyzing model output on the Rutgers University Office of Advanced Research Computing (OARC) cluster computer Amarel.
Amarel help
There is an OARC supported user guide at https://sites.google.com/view/cluster-user-guide to which you can refer for a lot of detailed guidance.
Working with UNIX
To work effectively on Amarel you need at least a beginner level of competency with the UNIX operating system. If you are a total newcomer to UNIX computers you might find it useful to work through a structured tutorial; for example Ryan's Linux Tutorial.
| Command | What it does | Example |
|---|---|---|
| ls | list contents of a directory files ending in .nc long format list and hidden files |
ls ls *.nc ls -al |
| cd | change directory | cd /projects/dmcs_1/courses/cod |
| pwd | print working directory to see where you are | pwd |
| cp | copy a file | cp source_file target_file |
| mv | rename a file move file to new directory |
mv file_name new_name mv file_name dir1/dir2/new_name |
| rm | delete (remove) a file | rm filename |
| more | type contents of a file to screen one page at a time | more .bashrc |
| less | like more only with more features (unix geeks are so funny) Read the man page for help |
less .bashrc |
| man | get the manual page (help) for a command | man less |
| . | the directory you are in | cp /projects/dmcs_1/courses/cod/job.sh . |
| .. | the directory above the one you are in | cd .. |
| grep | find a string in a file get regular expression and print (!) |
grep COD_1DMIX *.h |
Your default UNIX shell (the "flavor" of your UNIX environment) is bash, so for more advanced help it is often good to include "bash" as one of your search terms in a google search for help.
Editing files
One of the things that stumps newcomers to UNIX is making simple edits to files. Ryan's Linux Tutorial goes over the basics of the vi editor here, but it can be a beast to get comfortable with.
A more intuitive editor for new users is nano, the basics of which you can read about here.
There is also a way to edit files in the web browser based OnDemand service that is described later in this guide.
Rutgers VPN
If you are not on campus, you will need to connect to the Rutgers VPN. To use Rutgers VPN, you must first enroll in Duo 2 Factor Authentication (2FA) (NetID+). Most users have probably already done this. If not, navigate to https://netid.rutgers.edu/setupTwoFactorAuthentication.htm and follow the instructions.
Once you are enrolled in NetID+ you will need to activate the VPN service, if you have not already. Navigate to https://soc.rutgers.edu/vpn/ and click the gray button titled Service Activation. If you are not already logged in you may be asked to login and/or approve your login with the Duo Mobile app. To activate the VPN service, click the checkbox next to Remote Access VPN, Cisco AnyConnect Access for Rutgers and click the Activate Services button.
You are now ready to download the Cisco VPN client and connect to the Rutgers VPN. Complete instructions can be found here by clicking the red button titled General VPN Instructions to download the PDF. The instructions are geared towards Windows so if you are using a Mac you might find this page more helpful. In most cases, regardless of your operating system, pointing your browser to https://vpn.rutgers.edu/ and logging in with your NetID will lead you to downloading the correct VPN client.
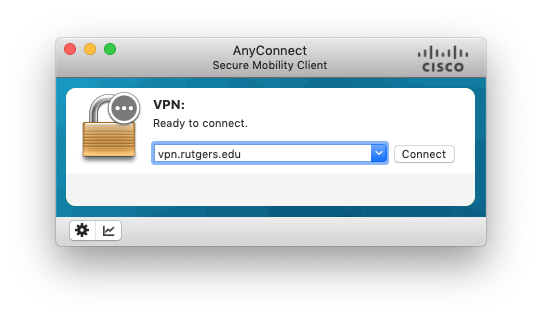
Once installed, open the Cisco AnyConnect client and type vpn.rutgers.edu in the box and click Connect:
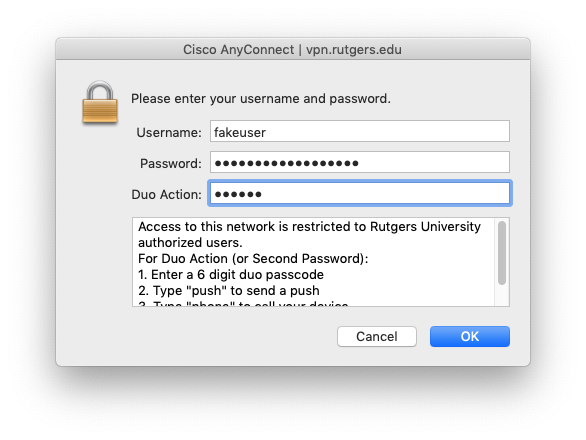
In the next window, shown below, Username will be your NetID.
In the second line Password is the password for your NetID.
The third line is the Duo Action to complete the 2FA, which can be accomplished in one of 4 possible ways:
- Enter a 6 digit Duo Passcode. These are generated either by a Hard Token, by showing the passcode in the Duo Mobile App, or from a previous “SMS” request. Simply type in the 6 numbers and hit OK.
- Type the word “push”. This will send a push notification to the primary device you have enrolled with Duo through NetID+ with the option to Accept or Deny.
- Type the word “phone”. You will receive a phone call to the primary device you have enrolled with Duo through NetID+ with touch tone options to Accept or Deny.
- Type the word “sms”. You will receive a text message to the primary device you have enrolled with Duo through NetID+ containing passcodes you can use to login.
Click OK and you should be connected to Rutgers VPN. You should have a small AnyConnect icon with a lock on it in the task tray (Windows ![]() ) or in the menu bar (Mac OS
) or in the menu bar (Mac OS ![]() ).
).
Connecting to Amarel with SSH
In order to connect to Amarel and compile and run ROMS you will need an SSH/terminal client. If you already use an SSH/terminal application you are comfortable with then stick with that and adapt the instructions below accordingly. For these instructions to work you need to either be on campus or connected to the Rutgers VPN.
Mac OS
If you are on a Mac, open the Terminal app (found in the Applications -> Utilities folder) or iTerm app and type:
- ssh fakeuser@amarel.rutgers.edu
replacing fakeuser with your NetID. When it asks if you want to continue connecting enter yes, then enter your NetID password.
Having to type the above command and netid password every time you login is moderately tedious, but this username/password authentication can become annoying and time consuming when repeatedly using the scp command to copy files to Amarel. Passwordless access and file transfer can be enabled using SSH keys by following the instructions below, or for more detail see the OARC User Guide on this topic.
- If you are still connected to Amarel, disconnect by entering exit
- At the prompt for your local machine, enter nano ~/.ssh/config and paste the block below at the end of the file:Host amarelreplacing NetID with your own NetID. Save and exit (Ctrl+o Ctrl+x). This allows you to type ssh amarel to connect instead of ssh fakeruser@amarel.rutgers.edu
Hostname amarel.rutgers.edu
HostKeyAlias amarel
User NetID - Enter ls ~/.ssh. If there is a file listed called id_rsa.pub skip to 5
- Enter ssh-keygen -t rsa, then hit return to accept the default location, enter a passphase twice (or we recommend you just leave it blank twice for no passphrase)
- Copy the public portion of the key to Amarel by entering scp ~/.ssh/id_rsa.pub amarel:.
- SSH to Amarel (ssh amarel) and check if you already have a .ssh folder:[rjdave@amarel1 ~]$ ls -l .sshYou probably won't have all the files above but if you instead get the message
total 33
-rw------- 1 rjdave rjdave 1069 Sep 8 2017 authorized_keys
-rw-r--r-- 1 rjdave rjdave 434 Jun 4 2021 config
-rw-r--r-- 1 rjdave rjdave 5992 Dec 2 13:47 known_hostsls: cannot access .ssh: No such file or directorythen enter the following command:
mkdir .ssh && chmod 700 .ssh.
These two commands (combined on one line) will create the .ssh folder and set the required permissions. Setting private user-only permissions (chmod 700 ...) is important, because the system will not allow you to have your keys viewable by others users. - Once you have a .ssh folder with the proper permissions, enter the following commands
cat ~/id_rsa.pub >> ~/.ssh/authorized_keys && chmod 640 ~/.ssh/authorized_keys
then exit back to your local machine and execute ssh amarel again. It should no longer ask for your password.
Windows
For Windows, we recommend MobaXterm installer edition.
- Once installed open it, choose light or dark theme and click the Session button in the upper left.
- Choose SSH and enter amarel.rutgers.edu for Remote host, check the Specify username box and enter your NetID in the box and click OK.
- You will be asked to type your NetID password and then asked if you want to save the password. If you choose yes you will be asked to set a master password to encrypt all your saved passwords.
- In the future you can click Sessions (not Session) and select amarel.rutgers.edu and not have to enter your password.
Setting up your .bashrc
The default software setup by OARC on Amarel does not include everything needed to compile and run ROMS. Ocean Modeling Group computing specialist David Robertson has configured, and maintains and updates, a repository of what you need for ROMS and other models that will be automatically loaded for every login session once you add some lines to the login script (.bashrc, .cshrc, etc.) in your home directory. Log in to Amarel with Terminal, iTerm2 or MobaXterm and edit your login script as described below.
Unless otherwise requested your default shell will be bash and the following lines shown below in red and green should be added (with nano or your preferred editor) near the top of your .bashrc after the import of the global definitions as shown:
- # Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
ulimit -s unlimited
export MODULEPATH=/projects/dmcs_1/sw/modulefiles/Core:${MODULEPATH}
export SQUEUE_FORMAT="%.18i %.9P %.8j %.8u %.2t %.10M %.10l %.4C %.6D %R"
# Automatically load the roms module
module load roms
The line with ulimit allows you to take full advantage of computing resources (may be obsolete now but it won't hurt anything).
The first export line will tell Lmod where to find the modules that will load the custom software.
The second export line makes the squeue command (explained later) more useful.
The lines in green are optional and will load the roms module automatically every time you login to Amarel. They must be after the line that sets the MODULEPATH.
Once you have added these lines, log out and back in for them to take effect.
![]() Note: If you see the error ...
Note: If you see the error ...
- Lmod has detected the following error: The following module(s) are unknown: "roms"
Please check the spelling or version number. Also try "module spider ..."
It is also possible your cache file is out-of-date; it may help to try:
$ module --ignore_cache load "roms"
Also make sure that all modulefiles written in TCL start with the string #%Module
... then you may need to logout and login again so that .bashrc sets MODULEPATH as instructed above. If the error persists, you may not have been properly added to the dmcs_1 Linux group. To check if you are part of dmcs_1 type the command id and if dmcs_1 is not listed, let us know and we will get you added to the group.
Accessing the ROMS Source Code
Register as a ROMS user
If you are not already a ROMS user, you will need to fill out the registration form found here and wait for approval. Once approved, you will be able to checkout the ROMS source code. It is highly recommended that you use the GitHub myroms repository to obtain the ROMS source code. The Subversion (SVN) repository is being phased out and will no longer be available after December 31, 2024. If you manage your own software projects using GitHub, the concepts for software version control may already be familiar to you. Detailed directions can be found on the WikiROMS Git page.
Obtaining the ROMS source code
There is nothing special about obtaining the ROMS source code on Amarel. The git commands are the same as on any UNIX host. Navigate to the directory where you want your ROMS source code to reside and execute this git clone command:
- git clone https://github.com/myroms/roms.git <my_src_dir>
replacing <my_src_dir> with what you want the source code directory to be called. You can also omit <my_src_dir> and git will automatically name the folder roms.
It can take a couple of minutes to download all of ROMS, but you only do this once.
Many users simply run this command from their home directory and use myroms for <my_src_dir>, but the choice is yours depending on how you want to manage your Amarel space and how many ROMS projects you think you might end up working on.
Loading and Unloading Modules
Like many clusters, Amarel uses environment modules to load and unload software and configure the environment. Some commands you will find useful are:
- module help Display help message
module help <m1> Show help information for module <m1>
module available Show all modules currently available
module whatis <m1> Show brief information about module <m1>
module spider List all possible modules even if not currently available
module list List currently loaded modules
module load Load the specified module(s)
module unload Unload the specified module(s)
module swap <m1> <m2> Unload <m1> and then load <m2> (for switching versions of the same software)
module purge Unload all loaded modules
Loading the ROMS Module
If you did not add the optional lines from the Setting up your .bashrc section, you can easily set up your environment to compile and run ROMS by manually loading the roms module.
- module load roms
module list
Currently Loaded Modules:
1) intel/17.0.4 2) mvapich2/2.2 3) mct-roms/2.6.0 4) netcdf/4.6.2
5) esmf/8.0.0_nc4 6) parpack-roms/2.1 7) hdf5/1.10.4 8) roms/intel_nc4
Notice that loading the roms module will actually load 7 other modules.
This will setup your environment to use the Intel compiler so remember to set FORT to ifort in your build script.
![]() Note: If you see the error ...
Note: If you see the error ...
- Lmod has detected the following error: The following module(s) are unknown: "roms"
Please check the spelling or version number. Also try "module spider ..."
It is also possible your cache file is out-of-date; it may help to try:
$ module --ignore_cache load "roms"
Also make sure that all modulefiles written in TCL start with the string #%Module
... you may not have been properly added to the dmcs_1 Linux group. To check if you are part of dmcs_1 type the command id and if dmcs_1 is not listed, let us know and we will get you added to the group.
Your project/coursework working directory space
We recommend you create a subdirectory of your home directory in which to work, so you can keep each ROMS project separate and organized.
You might call this working directory CoastalDynamics, or Projects, or simply cod (for coastal ocean dynamics). You will be typing this a lot, so keep it simple.
To make a new directory called Projects, enter this command
- mkdir Projects
Change to this working directory with the command
- cd Projects
In instructions that follow, when we refer to your working directory we mean this workspace.
Configuring and Compiling ROMS
You will compile and run ROMS in your project/coursework working directory, so change to that directory first.
- cd Projects
Running the build script build_roms.sh is the recommended method for compiling ROMS on Amarel. For any given project you will likely only need to change build_roms.sh to reset ROMS_APPLICATION and MY_ROMS_SRC and make sure FORT is set to ifort.
![]() It is important to make sure USE_MY_LIBS is set to no or your compilation will fail.
It is important to make sure USE_MY_LIBS is set to no or your compilation will fail.
A template build_roms.sh script is in subdirectory ROMS/Bin of <my_src_dir> that you created when you downloaded the code. For example, if you created myroms in your home directory the copy command would be
- cp ~/myroms/ROMS/Bin/build_roms.sh .
Now open build_roms.sh with an editor (e.g. nano) and modify the line MY_ROMS_SRC to point to your choice for <my_src_dir>. So, you will have something like ...
- export MY_ROMS_SRC={$HOME}/myroms
if the name you chose for <my_src_dir> was myroms.
If this is your first time working with ROMS, a good starting place is to compile the default UPWELLING test case that is indicated in build_roms.sh with the default setting export ROMS_APPLICATION=UPWELLING.
To compile the UPWELLING example, you need to copy the configuration file upwelling.h to your working directory. This is in subdirectory ROMS/Include of your source code. For example, the command might be:
- cp ~/myroms/ROMS/Include/upwelling.h .
Once your build script is configured and you have upwelling.h in your working directory you can compile ROMS by typing:
- ./build_roms.sh -j 4
where the number after -j indicates the number of compute cores to use in parallel to execute the compilation. The greater the number, the faster it goes.
However, the login node you will be compiling on is shared for the entire Amarel system. If you use a number larger than 4, or omit it altogether (which says use all cores on the login node) your build might be terminated by an administrator. Be a considerate user and keep the number low.
If compilation is processing successfully you will see a lot of information typed to your screen as each component of the source code is compiled. It will take a minute or so to complete.
If compilation ends quickly with errors like this:
- $ ./build_roms.sh -j 4
rm -f -r core *.ipo /home/jwilkin/cod/Projects/Build_roms...
which: no mpif90 in (/usr/lpp/mmfs/bin:/usr/local/bin:/us...
cp -f /opt/intelsoft/serial/netcdf3/include/netcdf.mod /h...
cp: cannot stat ‘/opt/intelsofo... No such file or directory
make: *** No rule to make target netcdf.mod needed by ... Stop.
... this means you have not run the module load roms command. Go back to the notes on Amarel#Loading_the_ROMS_Module.
If compilation was successful, there will be a file named romsM that is the model executable. You can check for the file with this command:
- ls -l *
You should see:
- Build_roms build_roms.sh romsM upwelling.h
Build_roms is a folder that build_roms.sh created to hold all the temporary files that the FORTRAN compilation process makes to combine into the romsM executable file.
Running on the Amarel Compute Nodes
When you ssh to Amarel you are connected to one of the login nodes. These nodes are to be used for file editing, transfering output and input files to/from your local computer, and modest code compiling tasks and analysis, but not for compute intensive tasks. Here we explain how to connect to the compute nodes for these larger tasks.
Note: Consult the Amarel status page (https://oarc.rutgers.edu/amarel-system-status/) before scheduling a job. Amarel is down for maintenance monthly.
What partitions can you run on?
Access to Amarel resources is controlled by partitions. You can find out the partitions you have permission to access with the command
- sinfo -s
This will show the partitions that everyone has access to. See the cluster guide entry on partitions for an explanation of these.
If you belong to a research group that own a partition you should also see that listed, for example p_omg_1 for the Ocean Modeling Group. This is the partition you want to use most of the time as your job will not get pre-empted by other owner users.
Using the SLURM batch job scheduler
Amarel uses SLURM workload manager to schedule compute intensive tasks. The user configures a SLURM job script for each model run and submits this script with the sbatch command. The job script declares the resources required, such as number of CPUs for parallel jobs, maximum memory required, etc.
We have configured a simple template job script (for the ROMS UPWELLING example) that you can copy from /projects/dmcs_1/courses/cod/job.sh into your working directory.
The UNIX command to copy a file is cp source_file target_file. So, you would enter
- cp /projects/dmcs_1/courses/cod/job.sh .
The syntax here uses a dot (.) for the target_file, which causes the source_file (job.sh) to be copied to the present working directory without changing its name.
When typing this, or any, UNIX command, you can start typing just the beginning few characters of the path then press TAB to autocomplete the filename. If the completion is ambiguous, you can continue typing more characters and press TAB again. Pressing TAB twice in a row will show you the set of possible completions. This trick can save you a lot of tedious (mis)typing.
The contents of job.sh are shown below:
- #!/bin/bash
#SBATCH --partition=p_omg_1 # Partition. If you are not a member of OMG enter your own partition
#SBATCH --job-name=upwelling # Assign a short name to your job
#SBATCH --nodes=1 # Number of nodes you require
#SBATCH --ntasks=4 # Total number of tasks you'll launch
#SBATCH --ntasks-per-node=4 # Number of tasks you'll launch on each node
#SBATCH --cpus-per-task=1 # Cores per task (>1 if multithread tasks)
#SBATCH --mem=6400 # Real memory (RAM) required (MB) per node
#SBATCH --time=00-00:05 # Total run time limit (DD-HH:MM)
#SBATCH --output=out.%N.%j # STDOUT output file
#SBATCH --error=err.%N.%j # STDERR output file (optional but recommended)
#SBATCH --export=ALL # Export you current env to the job env
## It is important to have --mpi=pmi2 here or ROMS will not run
srun --mpi=pmi2 ./romsM roms_upwelling.in
Note: The maximum run time is 14-00:00 (14 days). It is tempting to set this to the maximum in order to avoid a job ending too early. (i.e. Slurm stopping a ROMS run before it is finished) However, if there is maintenance scheduled during that time period the job will not start until maintenance is complete. So a runtime as close as possible to the actual run time is recommended.
Requesting a specific class of compute node
For large jobs it makes sense to use all available cores on a node, and for low priority jobs to use older less fast nodes. You can do this by requesting a specific node number, but it is better to request a particular class of nodes so you don't unwittingly pick a node already in use. The classes available from oldest to newest nodes are:
- skylake (32 cores) (Phase I nodes) (nodes hal0116 through hal0120) Note: These will be reclaimed for general use in Feb 2024
- cascadelake (40 cores) (Phase II nodes) (nodes hal0159 through hal0163)
- icelake (64 cores) (Phase V.B nodes) (nodes hal0299 -- hal0302 and hal0346 -- hal0351)
Make the node class request in the SLURM script and choose the maximum number of cores to match, e.g.
- #SBATCH --constraint=cascadelake # Run on Phase II nodes (40 cores)
#SBATCH --ntasks=40 # Total number of tasks to launch (like -np to mpirun)
#SBATCH --ntasks-per-node=40 # Number of tasks to launch on each node
If you don't specifically request a class of nodes the SLURM scheduler will make its own choice on the most appropriate resources to use given your request. We don't know the basis of that choice.
If you request a class of node that is fully assigned to other users in your partition, your job will wait for the others users jobs to complete. So, coordinate with your colleagues if resource use is at a premium. You can use the squeue -p partition_name command to see what resources are being used by other users, e.g.
- $ squeue -p p_omg_1
JOBID PARTITION NAME USER ST TIME TIME_LIMIT CPUS NODES NODELIST(REASON)
26975473 p_omg_1 fwd levinj R 17:15:42 14-00:00:00 80 2 hal[0159,0162]
Running the ROMS Upwelling Example
You may have noticed that the srun command above includes a file named roms_upwelling.in. You will need to copy this file and varinfo.yaml from the ROMS/External directory of your <my_src_dir> ROMS source code to the directory where you compiled ROMS.
If, for example, you chose myroms as the name of your <my_src_dir> when you downloaded the code, you would enter
- cp ~/myroms/ROMS/External/roms_upwelling.in .
The syntax here uses tilde (~) as shorthand for your home directory,
After you copy the files, you will need to make a couple of small edits (using e.g. nano) to roms_upwelling.in to get this to work.
- Change the line with VARNAME to read VARNAME = varinfo.yaml (i.e. delete the ROMS/External part)
- Set NtileI == 2 and NtileJ == 2.
The product of NtileI and NtileJ, i.e. 2 x 2 = 4, is the number of cores the model will run on in parallel. This number must match the number in the SLURM job script options --ntasks and, in this case, --ntasks-per-node
Now you can submit your job to sbatch with the command sbatch job.sh
ROMS configurations for Coastal Ocean Dynamics class
The files that will configure the examples we will study in class will be located in /projects/dmcs_1/courses/cod.
You will find instructions for configuring and running the examples at a separate Wiki page COD_Class_Examples
Checking Job Status with squeue
Detailed documentation for monitoring your SLURM jobs can be found here. The easiest way to check whether your job is running is with the command squeue -p p_omg_1 (or your own partition name if it is not p_omg_1).
You should see output something like this:
- JOBID PARTITION NAME USER ST TIME TIME_LIMIT CPUS NODES NODELIST(REASON)
17197219 dmcs_1 watl_psas rjdave R 3-04:07:00 3-20:00:00 4 1 hal0035
17146609 dmcs_1 upwelling rjdave R 00:01:10 00:05:00 4 1 hal0035
You can see that the job is running (the ‘R’ under ‘ST’), has been running for 1 minute, 10 seconds, and is running on node 35.
Checking Progress with tail
You can check ROMS progress by using the tail command on the output file. For the upwelling job above the file would be called out.hal0035.17146609 so the command tail out.hal0035.17146609 will show you the most recent 10 lines written to the output log and will most likely tell you what time-step the model is on. Using the -f option (tail -f out.hal0035.17146609) will output appended data as the file grows. Ctrl-C will escape the display.
Did my UPWELLING job run?
If the UPWELLING test case was successful there should be an output log file named something like out.hal0035.17146609 that shows no errors, and there should be 4 output NetCDF files ...
- ls -l *.nc
roms_avg.nc
roms_dia.nc
roms_his.nc
roms_rst.nc
Cancelling a Job
To safest way cancel a queued or running job is to use its jobid. A job can be canceled by name but that is not recommended. To cancel the upwelling job in the example above you would issue the command scancel 17146609. If the job has not yet started, it will be removed from the queue. If it is running, all child processes will be killed and the job will be removed from the queue. You are only able cancel jobs that you own.
Running an interactive session on a compute node
It is possible to conduct your work interactively on one of the compute nodes (instead of the login node). For most work we will be doing in class this is not necessary, but if for some reason you have a job that needs many processors or a large amount of memory, and you want to run it interactively - say, to simply check that everything is in order for it to start correctly - there are instructions in the Cluster User Guide here.
For compute intensive interactive work, such as model analysis using Python or MATLAB, we recommend using the OnDemand interface to launch an interactive session on a set of compute nodes.
Using the OnDemand desktop environment
The OnDemand service allows you to connect to Amarel using a graphical desktop environment in a web browser. One of the most useful features of OnDemand is that it allows you to analyze model output by Python or Matlab on the cluster compute nodes.
You need to be running a Rutgers VPN to use OnDemand.
Navigate your browser to https://ondemand.hpc.rutgers.edu/pun/sys/dashboard and login with your NetID and password. At the top of the page click Interactive Apps.
Python Jupyter notebook:
- Select Personal Jupyter from the list on the left
- Make your selection for number of hours your session can remain active (e.g. 3) hours
- Set the number of cores to run on (I suggest 1 for now)
- Set memory (8 GB is an OK choice)
- For Partition enter e.g. p_omg_1
- Leave Reservation blank
- For Slurm feature enter cascadelake so your session starts on our older nodes and does not interfere take the larger, faster cores being used by OMG
- For conda path enter /projects/dmcs_1/miniconda3
- For conda environment enter /projects/dmcs_1/sw/packages/xroms/py38. This conda environment built for DMCS_1 makes available many of the Python modules you are used to using.
These settings are remembered the next time you access OnDemand.
Click Launch and wait. This can take a couple minutes.
You should see sub-window of the web page like this
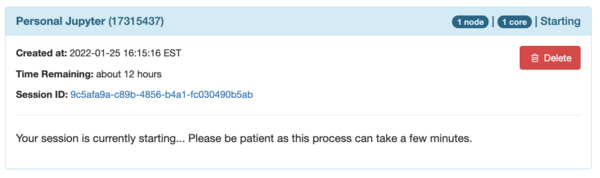
When the session is ready it changes to this:
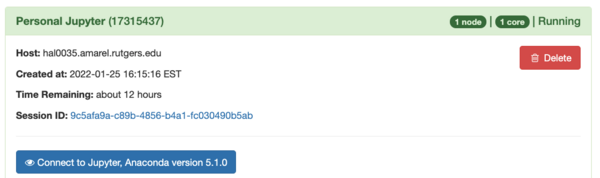
Start your notebook by clicking the Connect to Jupyter, Anaconda version 5.1.0 button: (If this does not appear, check that you correctly entered the conda path and conda environment at step 3 above.) 
This should place you in a file browse window from which you can navigate to one of your existing Jupyter notebooks, or, to start a new Untitled notebook, select Python 3 (ipykernel) from the New pull-down menu on the right side of the page 
Once your notebook is running, you can import the xroms package as you would any other toolbox, by entering import xroms then hold down the shift key and hit the return/enter key. You will see an asterisk [*] in the square brackets.
At present the import steps are quite slow ... 30 seconds ... so be patient and don't re-run those cells in your notebook if you don't have to.
Matlab:
- Select MATLAB from the list on the left
- As for Step 2 above for Python, choose time, cores, and memory
- Select MATLAB version, and enter p_omg_1 (or your own group name) in partition.
- Click Launch and wait. This can take a couple minutes.
- Once the Launch noVNC in New Tab button appears, click it and a MatLab GUI will open.
You are running in a graphical desktop environment, so by default Matlab will look in Documents/MATLAB/startup.m for your personal Matlab configuration file.
Popular MATLAB utilities for working with ROMS output are in /projects/dmcs_1/sw/packages/matlab-tools. These include the myroms.org utilities in subdirectory roms_matlab, and John Wilkin's toolbox in roms_wilkin.
Reconnecting to an interactive session
A particularly nifty feature of OnDemand is that your interactive session is independent of the browser window instance.
Say you are working on your office computer using a Jupyter notebook via OnDemand but haven't finished all you want to do by the end of the day. If you leave that session running, you can go home and connect once again to OnDemand and you will see your session in the list of Interactive Sessions. Launch that session and you will be placed right back where you were. OnDemand is simply a web interface to code running on Amarel, so you can connect from any browser.
Of course, this only works if you selected a number of hours in Step 2 above that will allow your session to remain open.
As long as the session is active the Amarel resources (cores and memory) are reserved for you and no one else. So, be respectful of other users and close out an interactive session once you are done.