Custom Query (986 matches)
Results (724 - 726 of 986)
| Ticket | Owner | Reporter | Resolution | Summary |
|---|---|---|---|---|
| #864 | Fixed | IMPORTANT: Corrected bug in LwSrc | ||
| Description |
In src:ticket:860, an update to vertical influx point sources (like river runoff) was released. However, the IF-conditional in step3d_t.F was outside of the tracer DO-loop. We need to have instead: IF (LwSrc(ng)) THEN
DO itrc=1,NT(ng)
IF (.not.((Hadvection(itrc,ng)%MPDATA).and. &
& (Vadvection(itrc,ng)%MPDATA))) THEN
DO is=1,Nsrc(ng)
Isrc=SOURCES(ng)%Isrc(is)
Jsrc=SOURCES(ng)%Jsrc(is)
IF (((Istr.le.Isrc).and.(Isrc.le.Iend+1)).and. &
& ((Jstr.le.Jsrc).and.(Jsrc.le.Jend+1))) THEN
DO k=1,N(ng)
cff=dt(ng)*pm(i,j)*pn(i,j)
# ifdef SPLINES_VDIFF
cff=cff*oHz(Isrc,Jsrc,k)
# endif
IF (LtracerSrc(itrc,ng)) THEN
cff3=SOURCES(ng)%Tsrc(is,k,itrc)
ELSE
cff3=t(Isrc,Jsrc,k,3,itrc)
END IF
t(Isrc,Jsrc,k,nnew,itrc)=t(Isrc,Jsrc,k,nnew,itrc)+ &
& cff*SOURCES(ng)%Qsrc(is,k)* &
& cff3
END DO
END IF
END DO
END IF
END DO
END IF
The TLM, RPM, ADM version of step3d_t.F was also updated. Many thanks to Chuning Wang and John Wilkin for reporting this bug. |
|||
| #865 | Done | IMPORTANT: Split 4D-Var algorithms, Phase II | ||
| Description |
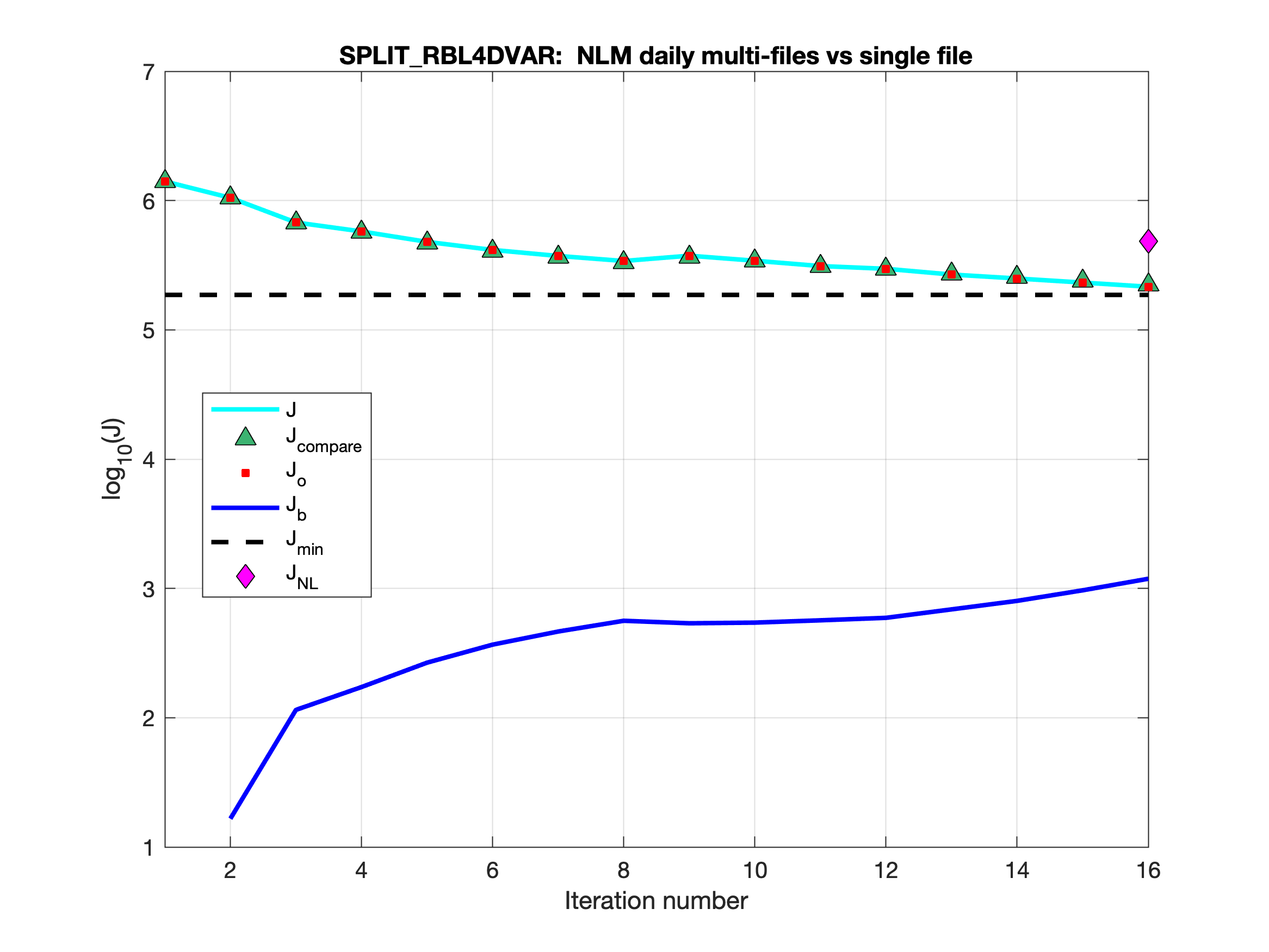
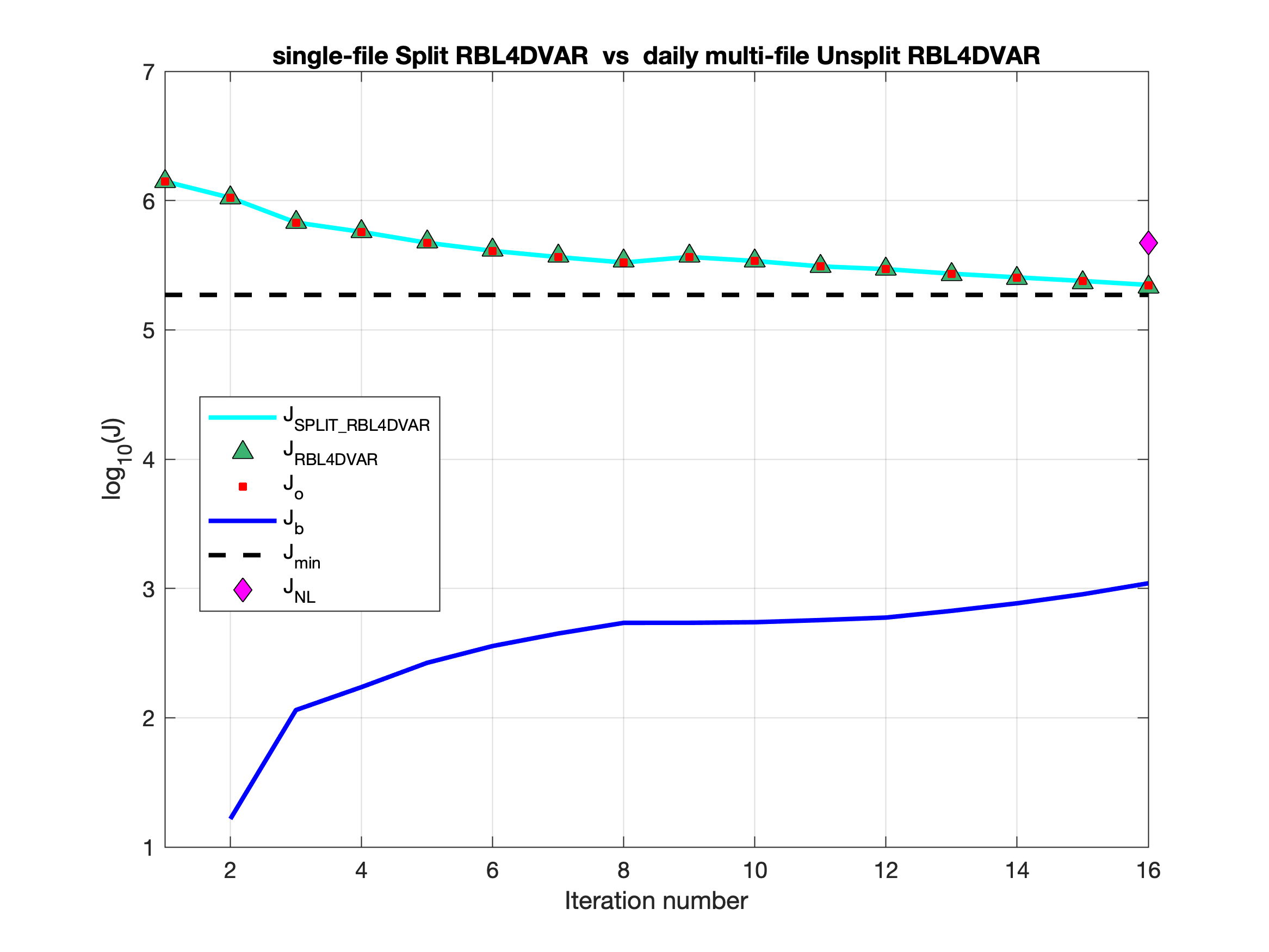
Testing is completed for the basic split 4D-Var algorithms: I4DVAR_SPLIT, RBL4DVAR_SPLIT, and R4DVAR_SPLIT. Now, we get identical solutions when comparing unsplit and split solutions with the WC13 test case. The split strategy is scripted to run with different executables in the elementary background, increment, and analysis 4D-Var phases, as mentioned in src:ticket:862. The modules i4dvar.F, rbl4dvar.F, and r4dvar.F have additional code activated with the internal CPP option SPLIT_4DVAR to load values to several variables which are assigned independently in other phases or are in memory in the unsplit algorithm.
Corrected Bug:Corrected a multi-file bug in output.F, tl_output.F, rp_output.F, and ad_output.F for all ROMS output files in restart applications. For example in output.F, the counter HIS(ng)%load needs reset to zero in restart applications: IF ((nrrec(ng).ne.0).and.(iic(ng).eq.ntstart(ng))) THEN
IF ((iic(ng)-1).eq.idefHIS(ng)) THEN
HIS(ng)%load=0 ! restart, reset counter
Ldefine=.FALSE. ! finished file, delay
ELSE ! creation of next file
Ldefine=.TRUE.
NewFile=.FALSE. ! unfinished file, inquire
END IF ! content for appending
idefHIS(ng)=idefHIS(ng)+nHIS(ng) ! restart offset
ELSE IF ((iic(ng)-1).eq.idefHIS(ng)) THEN
...
END IF
The same needs to be corrected for the AVG(ng), ADM(ng), QCK(ng), and TLM(ng) structures field load. Many thanks to Julia Levin for bringing this to my attention. |
|||
| #866 | Fixed | Missing Drivers directory makefile macros | ||
| Description |
The makefile in the git repository needs be modified in order to compile Drivers sub-directotory library: modules += ROMS/Nonlinear \
ROMS/Nonlinear/Biology \
ROMS/Nonlinear/Sediment \
ROMS/Functionals \
ROMS/Drivers \
ROMS/Utility \
ROMS/Modules
The WC13 I4d-Var gives the following error during compilation: libDRIVER.a(i4dvar.o): In function `i4dvar_mod_mp_background_': i4dvar.f90:(.text+0x60f): undefined reference to `initial_' i4dvar.f90:(.text+0x90d): undefined reference to `main3d_' libDRIVER.a(i4dvar.o): In function `i4dvar_mod_mp_increment_': i4dvar.f90:(.text+0x1977): undefined reference to `tl_main3d_' i4dvar.f90:(.text+0x19c3): undefined reference to `ad_initial_' i4dvar.f90:(.text+0x1a4b): undefined reference to `tl_initial_' i4dvar.f90:(.text+0x1c99): undefined reference to `ad_main3d_' i4dvar.f90:(.text+0x1d9d): undefined reference to `ad_variability_mod_mp_ad_variability_' i4dvar.f90:(.text+0x1dba): undefined reference to `ad_convolution_mod_mp_ad_convolution_' i4dvar.f90:(.text+0x24ee): undefined reference to `tl_convolution_mod_mp_tl_convolution_' i4dvar.f90:(.text+0x2507): undefined reference to `tl_variability_mod_mp_tl_variability_' i4dvar.f90:(.text+0x2660): undefined reference to `tl_wrt_ini_' i4dvar.f90:(.text+0x275a): undefined reference to `ad_wrt_his_' i4dvar.f90:(.text+0x3140): undefined reference to `tl_wrt_ini_' i4dvar.f90:(.text+0x3f0a): undefined reference to `tl_wrt_ini_' i4dvar.f90:(.text+0x3f46): undefined reference to `tl_wrt_ini_' i4dvar.f90:(.text+0x3f82): undefined reference to `tl_wrt_ini_' |
|||