| Description |
The code in lines 659–714 of ROMS/include/globaldefs.h defines the symbol FRC_FILE. Then in lines 5576–5589 of ROMS/Utility/read_phypar.F the model checks for the existence of the file(s) specified by the FRCNAME entry in ocean.in. The globaldefs.h code defines FRC_FILE if, amongst other conditions, either SSH_TIDES or UV_TIDES is set: see line 700 if SOLVE3D is set and line 711 if it is not.
However, tidal parameters are now read from special tidal-forcing files (entry TIDENAME), not from ordinary forcing files as they were previously. The current code fails when tides are required but with no other forcings.
The references in ROMS/include/globaldefs.h to
(defined SSH_TIDES || defined UV_TIDES)
in lines 700 & 711 should be removed.
|
| Description |
Added new vertical stretching option, Vstretching=5, based on a quadratic Legendre polynomial function that allows higher resolution near the surface developed by Souza et al. (2015). Many thanks to Joao Sousa and Brian Powell for coding and testing this new vertical stretching. For more documentation, please check WikiROMS or the publication reference.
- The new vertical stretching functional is coded in set_scoord.F:
!
!----------------------------------------------------------------------
! Stretching 5 case using a quadratic Legendre polynomial function
! aproach for the s-coordinate to enhance the surface exchange layer.
!
! J. Souza, B.S. Powell, A.C. Castillo-Trujillo, and P. Flament, 2015:
! The Vorticity Balance of the Ocean Surface in Hawaii from a
! Regional Reanalysis.'' J. Phys. Oceanogr., 45, 424-440.
!
! Added by Joao Marcos Souza - SOEST - 05/07/2012.
!-----------------------------------------------------------------------
!
ELSE IF (Vstretching(ng).eq.5) THEN
SCALARS(ng)%sc_w(N(ng))=0.0_r8
SCALARS(ng)%Cs_w(N(ng))=0.0_r8
DO k=N(ng)-1,1,-1
rk=REAL(k,r8)
rN=REAL(N(ng),r8)
sc_w=-(rk*rk - 2.0_r8*rk*rN + rk + rN*rN - rN)/(rN*rN - rN)- &
0.01_r8*(rk*rk - rk*rN)/(1.0_r8 - rN)
SCALARS(ng)%sc_w(k)=sc_w
IF (theta_s(ng).gt.0.0_r8) THEN
Csur=(1.0_r8-COSH(theta_s(ng)*sc_w))/ &
& (COSH(theta_s(ng))-1.0_r8)
ELSE
Csur=-sc_w**2
END IF
IF (theta_b(ng).gt.0.0_r8) THEN
Cbot=(EXP(theta_b(ng)*Csur)-1.0_r8)/ &
& (1.0_r8-EXP(-theta_b(ng)))
SCALARS(ng)%Cs_w(k)=Cbot
ELSE
SCALARS(ng)%Cs_w(k)=Csur
END IF
END DO
SCALARS(ng)%sc_w(0)=-1.0_r8
SCALARS(ng)%Cs_w(0)=-1.0_r8
!
DO k=1,N(ng)
rk=REAL(k,r8)-0.5_r8
rN=REAL(N(ng),r8)
sc_r=-(rk*rk - 2.0_r8*rk*rN + rk + rN*rN - rN)/(rN*rN - rN)- &
0.01_r8*(rk*rk - rk*rN)/(1.0_r8 - rN)
SCALARS(ng)%sc_r(k)=sc_r
IF (theta_s(ng).gt.0.0_r8) THEN
Csur=(1.0_r8-COSH(theta_s(ng)*sc_r))/ &
& (COSH(theta_s(ng))-1.0_r8)
ELSE
Csur=-sc_r**2
END IF
IF (theta_b(ng).gt.0.0_r8) THEN
Cbot=(EXP(theta_b(ng)*Csur)-1.0_r8)/ &
& (1.0_r8-EXP(-theta_b(ng)))
SCALARS(ng)%Cs_r(k)=Cbot
ELSE
SCALARS(ng)%Cs_r(k)=Csur
END IF
END DO
END IF
The new functional is similar to Vstretching=4, but with a quadratic instead or linearly varying fractional vertical coordinate, s.
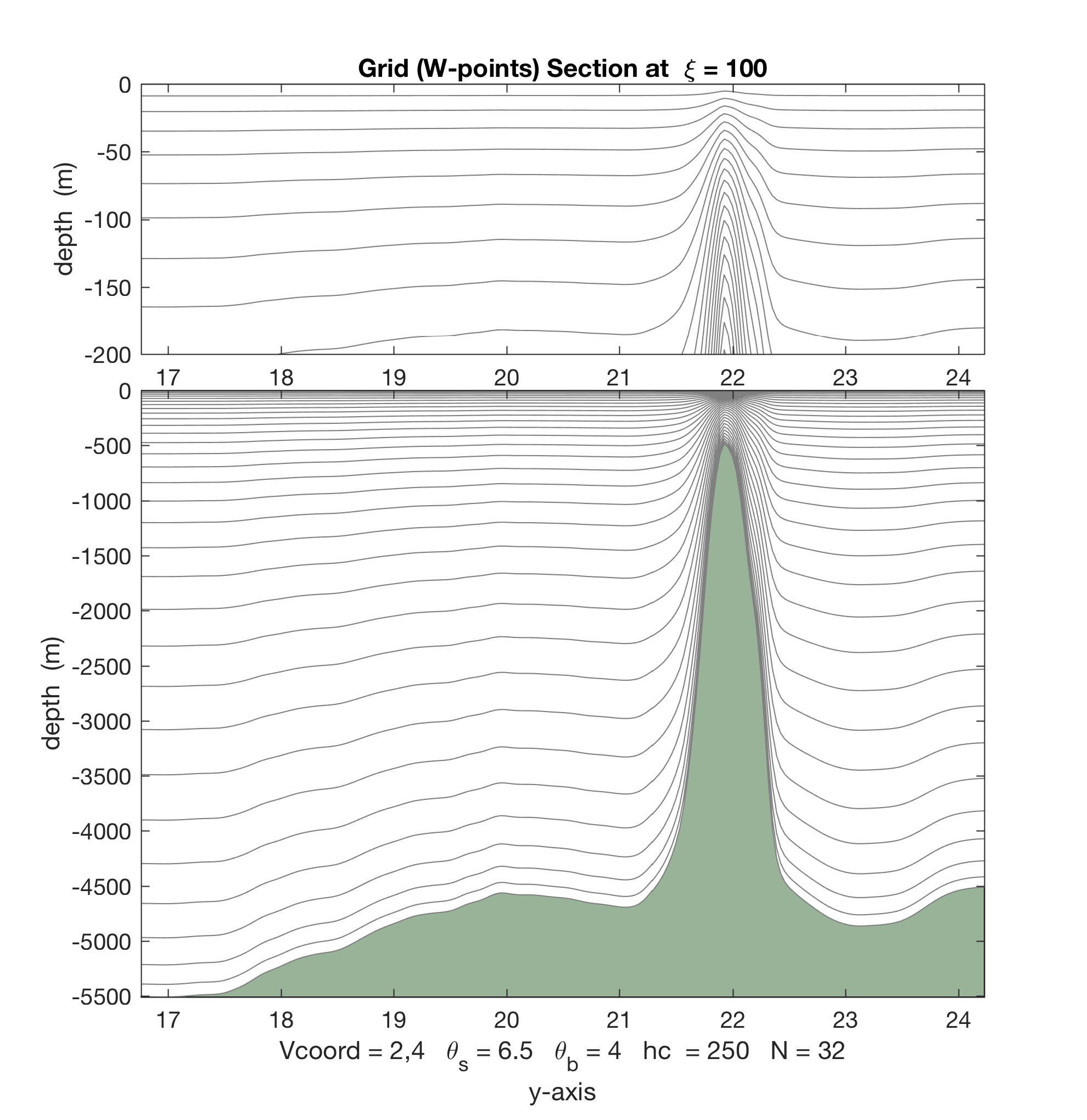
- The Matlab scripts stretching.m, set_depth.m, and scoord.m were updated with the new vertical stretching option. The difference between Vstretchin=4 and Vstretching=5 is illustrated below.
For Vtransform=2 and Vstretching=4, we get almost 4 vertical levels in the top 50 m:
Contrarily, with Vtransform=2 and Vstretching=5, we double the number of vertical levels to 8 in the top 50 m:
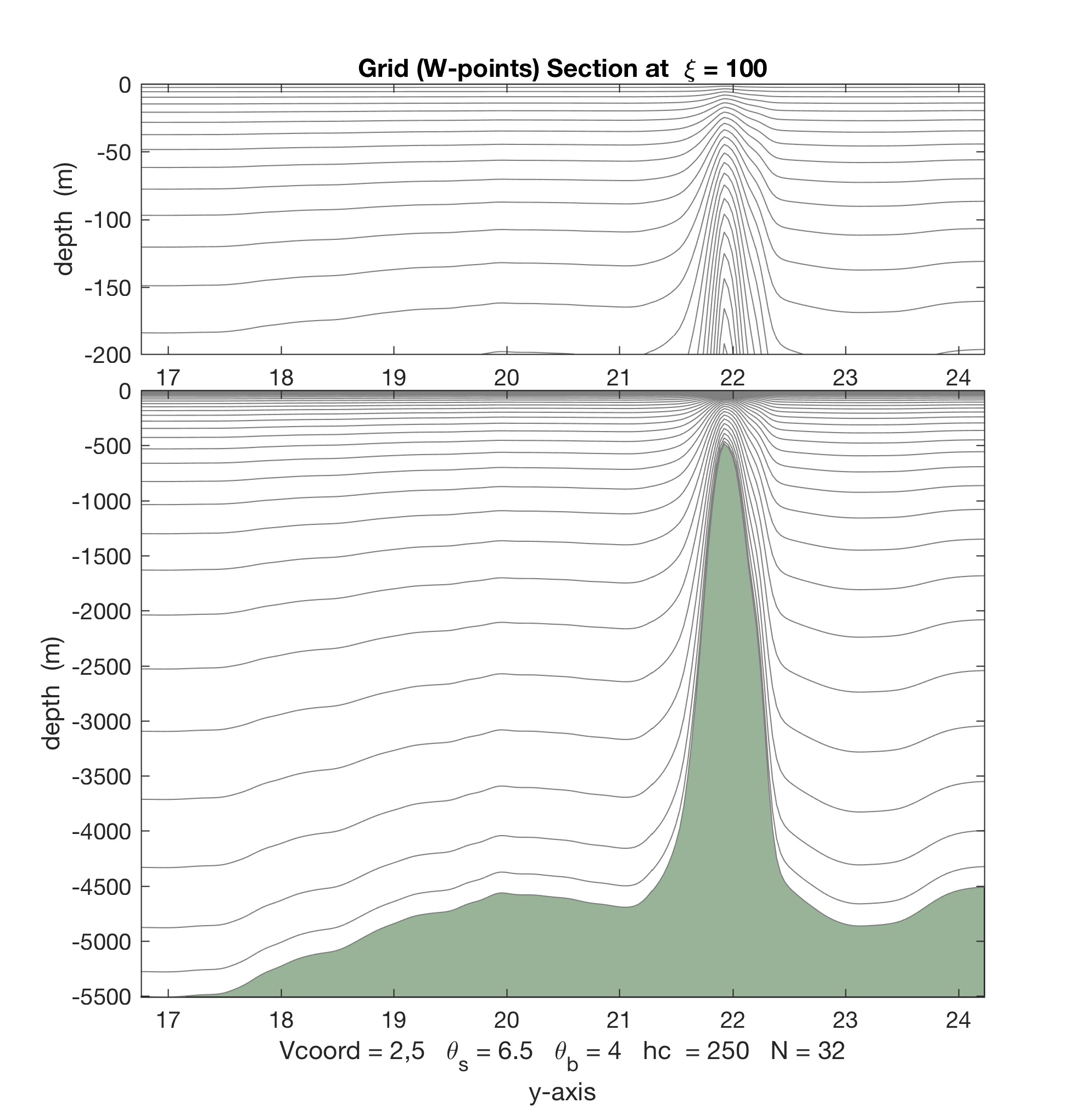
Notice that the increased vertical resolution at the near-surface causes a loss of resolution at the bottom.
- All the input ocean_*.in scripts were modified to update the documentation about Vstretching.
Reference:
Souza, J. M., B. Powell, A. C. Castillo-Trujillo, and P. Flament, 2015: The Vorticity Balance of the Ocean Surface in Hawaii from a Regional Reanalysis, J. Phys. Oceanogr., 45, 424-440, doi:10.1175/JPO-D-14-0074.1.
|
| Description |
- Corrected a bug in ad_misfit.F and obs_write.F in the managing of the screening and quality control variable obs_scale for the 4DVAR primal formulation. The issue is that in the primal formulation (IS4DVAR), the current time window (or survey) observations are loaded to the working arrays using local indices (array elements 1:Nobs) as opposed to global indices in the dual formulation (W4DPSAS and W4DVAR). Recall that the screening variable ObsScale is computed only once to facilitate Background Quality Control on the first pass of the NLM (WrtObsScale=T and wrtNLmod=T). Therefore, we need to load and save values into global array ObsScaleGlobal so it can be used correctly by the TLM, RPM, and ADM.
# ifndef WEAK_CONSTRAINT
!
!-----------------------------------------------------------------------
! Save observation reject/accept flag in GLOBAL screening variable.
!-----------------------------------------------------------------------
!
IF (wrtObsScale(ng).and.wrtNLmod(ng)) THEN
ic=0
DO iobs=NstrObs(ng),NendObs(ng)
ic=ic+1
ObsScaleGlobal(iobs)=ObsScale(ic)
END DO
ELSE
ic=0
DO iobs=NstrObs(ng),NendObs(ng)
ic=ic+1
ObsScale(ic)=ObsScaleGlobal(iobs)
END DO
END IF
# endif
Then, in ad_misfit.F, we just need to load the correct ObsScale for the current time assimilation window:
!
!-----------------------------------------------------------------------
! Load observation reject and accept flag into screening variable.
!-----------------------------------------------------------------------
!
ic=0
DO iobs=NstrObs(ng),NendObs(ng)
ic=ic+1
ObsScale(ic)=ObsScaleGlobal(iobs)
END DO
- Updated input 4DVAR script s4dvar.in to include the extra-observation parameters:
! In any application, the number of observation types is computed as:
! NobsVar = NstateVar + NextraObs.
!
! NextraObs values are expected for keywords ExtraIndex and ExtraName.
! If NextraVar > 1, enter one observation type names per line and
! use a backslash as the continuation.
NextraObs = 0
ExtraIndex = 20
ExtraName = radial
Here, NextraObs is the number of extra-observation classes to consider in addition to those associated with the state variables (one-to-one correspondence). They are used in observation operators that require more that one state variable to evaluate a particular extra-observation type like HF radials, travel time, pressure, or any other complex observation operator to be implemented in the future. In any application, the number of observation types is now computed as:
NobsVar(ng) = NstateVar(ng) + NextraObs
If not processing extra-observation classes, set NextraObs=0.
The parameter ExtraIndex defines the extra-observation class identification indices, as specified in the input observation NetCDF file variable obs_type. The index has to be unique and a number greater than 7+2*NT, where NT is the total of active plus passive tracers. The 7 values include zeta, ubar, vbar, u, v, sustr, and svstr. The 2 factor for NT is to account for the surface tracer flux. Therefore, avoid using 1:7+2*NT numbers for indices because they are reserved for the state vector. Be aware that you may want to use biological data assimilation in the future. NextraObs values are expected for this keyword. This parameter is only processed when NextraObs > 0.
The keyword ExtraName defines the extra-observation class names. NextraObs values are expected. This parameter is only processed when NextraObs > 0. Enter one type name per line and use a backslash for continuation. For example:
ExtraName = radial \
pressure
Currently, we are exploring the implementation of complex observation operators. This update is a structural change to facilitate implementing such operators in the future.
- Introduced mapping indices from state variable to observation type (ObsState2Type) and its inverse from observation type to state variable (ObsType2State). It is done because the extra-observation index (ExtraIndex) may not be in sequential enumeration with respect to regular observation types.
In obs_write.F, the ObsState2Type is used as follows:
IF (wrtNLmod(ng).and. &
& (FOURDVAR(ng)%ObsCount(isFsur).gt.0)) THEN
CALL extract_obs2d (ng, 0, Lm(ng)+1, 0, Mm(ng)+1, &
& LBi, UBi, LBj, UBj, &
& ObsState2Type(isFsur), & !<==
& Mobs, Mstr, Mend, &
& rXmin(ng), rXmax(ng), &
& rYmin(ng), rYmax(ng), &
& time(ng), dt(ng), &
& ObsType, ObsVetting, &
& Tobs, Xobs, Yobs, &
& OCEAN(ng)%zeta(:,:,KOUT), &
# ifdef MASKING
& GRID(ng)%rmask, &
# endif
& NLmodVal)
In obs_cost.F, the reverse mapping ObsType2State is used as follows:
DO iobs=NstrObs(ng),NendObs(ng)
ivar=ObsType2State(ObsType(iobs)) !<==
IF ((ivar.gt.0).and.(ObsScale(iobs).gt.0.0_r8).and. &
& (ObsErr(iobs).ne.0.0_r8)) THEN
cff=ObsScale(iobs)*(NLmodVal(iobs)-ObsVal(iobs))**2/ &
& ObsErr(iobs)
my_ObsCost(0)=my_ObsCost(0)+cff
my_ObsCost(ivar)=my_ObsCost(ivar)+cff
END IF
END DO
|


