Custom Query (986 matches)
Results (613 - 615 of 986)
| Ticket | Owner | Reporter | Resolution | Summary |
|---|---|---|---|---|
| #739 | Done | Updated the Red Tide model | ||
| Description |
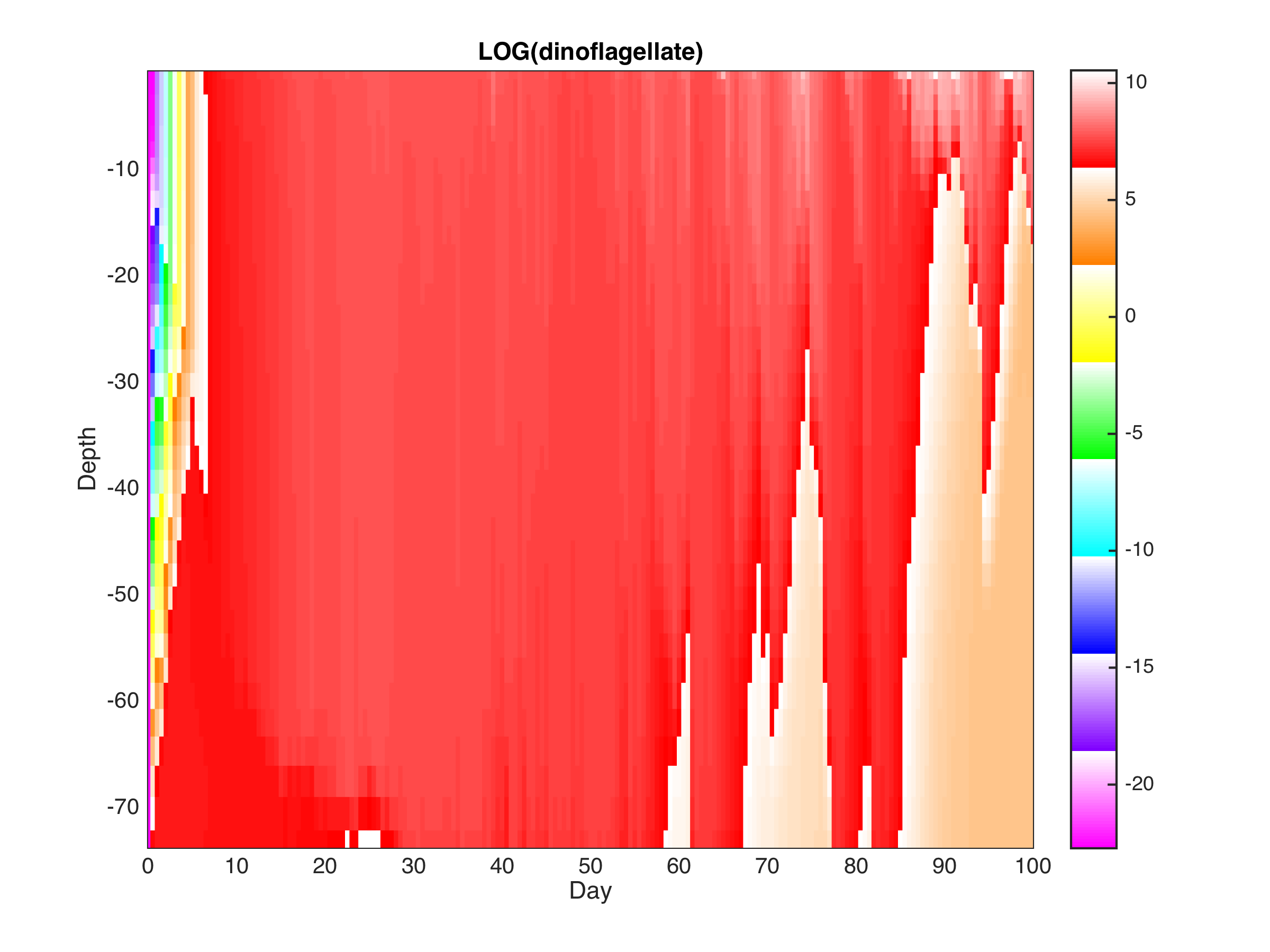
The red tide model (RED_TIDE) was updated to include a Q10 mortality rate equation, as a function of temperature. It includes new input parameters in red_tide.in: ! Mortality rate Q10 equation parameters, as a function of temperature:
!
! M_rate = Mor_a * Mor_Q10^[(temperature-MOR_To)/10] + Mor_b
MOR_a == 0.04d0 ! amplitude, 1/day
MOR_b == 0.03d0 ! intercept, 1/day
MOR_Q10 == 10.0d0 ! reaction rate base
MOR_T0 == 15.0d0 ! background temperature (C)
A couple of input parameters are also adjusted in red_tide.in for the Gulf of Maine:
! Maximum growth rate at optimal temperature and salinity [1/day]
Gmax == 1.05d0 ! Gmax
! Germination depth [cm]
Dg == 0.18d0 ! Dg
The BIO_TOY example gives the following Alexandrium Fundyense concentration: Notice that I am plotting the LOG of the concentration. Many thanks to Yizhen Li for the testing and update of the Red Tide Model. |
|||
| #742 | Fixed | Faulty proprocessor logic for DIAGNOSTICS_BIO | ||
| Description |
In ROMS/Include/globaldefs.h at lines 765-768 the effect of the following lines... #if (!defined BIO_FENNEL && defined DIAGNOSTICS_BIO) || \
(!defined HYPOXIA_SRM && defined DIAGNOSTICS_BIO)
# undef DIAGNOSTICS_BIO
#endif
...is to undefine DIAGNOSTICS_BIO unless both BIO_FENNEL and HYPOXIA_SRM are defined. I suggest... #if defined DIAGNOSTICS_BIO && !(defined BIO_FENNEL || HYPOXIA_SRM) # undef DIAGNOSTICS_BIO #endif ...or its syllogistic equivalent. Oops, the defined is missing before HYPOXIA_SRM. We need to have instead: #if defined DIAGNOSTICS_BIO && \ !(defined BIO_FENNEL || defined HYPOXIA_SRM) # undef DIAGNOSTICS_BIO #endif |
|||
| #743 | Fixed | Incomplete pre-processer conditions in def_diags.F | ||
| Description |
I think, in lines 626 and 701 of def_diags.F the pre-processor conditions should read as: #if defined DIAGNOSTICS_BIO && defined BIO_FENNEL Currently, they only read as: #ifdef BIO_FENNEL This can cause "undefined variable" errors for iDbio2 and iDbio3 during compilation when other diagnostics but not DIAGNOSTICS_BIO are enabled in combination with BIO_FENNEL. Yes, thank you for bringing this problem to my attention. I cleaned the logic in def_diags.F, set_diags.F, and wrt_diags.F. I also added the 2D biological diagnotics for HYPOXIA_SRM. It was not included in the above files. Also removed apostrophes in fennel.h documentation to suppress the warnings from ifort during compilation. I also cleaned a DIAGNOSTIC_BIO bug in hypoxia_srm.h. We don't need the 3D diagnostics arrays in this simple model. Fixed OMP directives in initial.F, ad_initial.F, rp_initial.F, and tl_initial.F when the 4D-Var algorithms are activated. This bug is of no consequence since we cannot run any of the adjoint-based algorithms in shared-memory (OpenMP). |
|||