| Description |
- The Matlab script contact.m was updated to compute the interpolation weights correctly when there is land/sea masking points in the contact regions. A new local function, interp_weights, is added to compute the weights at the contact points within the donor grid cell:
index4 index3
4______________3 (Idg+1,Jdg+1)
| : |
| : p |
| 1-q <----->|
| : |
Jr|..:....x : |
| . : |
| 1-p . : |
|<------> q |
| . : |
| . : |
|___________:__|
(Idg,Jdg) 1 Ir 2
index1 index2
For linear interpolation and all water points, the weights are:
W(1,:) = (1 - p) * (1 - q)
W(2,:) = p * (1 - q)
W(3,:) = p * q
W(4,:) = (1 - p) * q
If any of the donor grid points is masked, the associated weight is
set to zero and the other weights are re-scaled accordingly.
The following figure shows a two-way nesting application in the South China Sea with a 1:3 refinement grid over the Luzon Straits. Both grids are rotated. This is a tidally forced application. Tides are imposed at the coarser, large scale grid open boundaries.The figure shows both coarser and finer grids free-surface fields after 10 days of solution. Notice the train of internal waves generated at Luzon Straits. It is very hard to see where is the finer grid in this application so I have to draw the perimeters (red outline). The two-way nesting is perfect between the coarser and finer grids... We are very happy with this configuration and solution. Many thanks to John Wilkin for his help with this application.
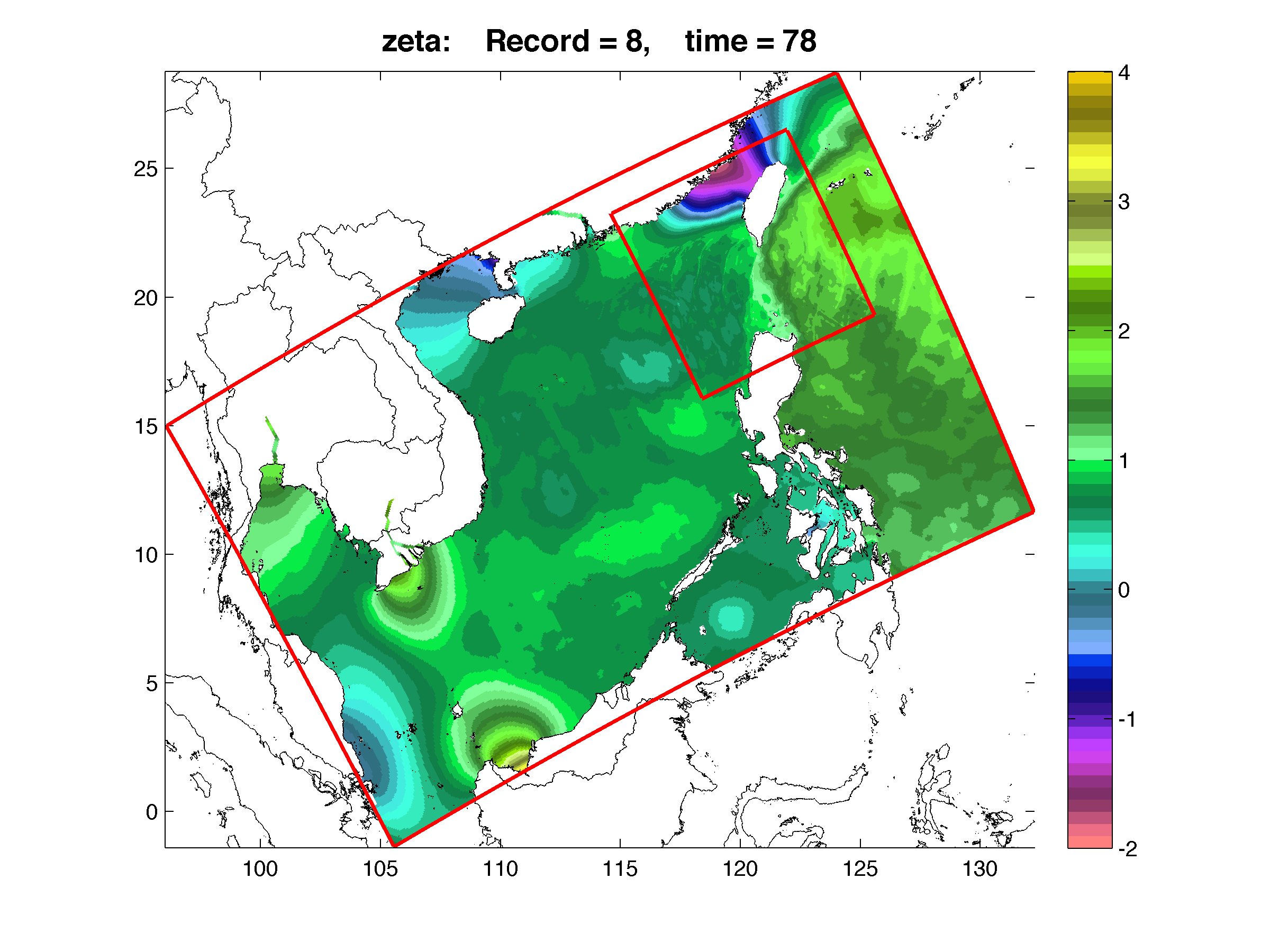
WARNING: You will need to regenerate the contact points NetCDF file using this updated version of contact.m if the donor grid has land/sea masking points in the contact region!!! This is the case in the above application. There is land/sea making in the southern and northern boundaries of the Luzon Straits refinement grid.
I also added few Matlab scripts:
- matlab/utility/pcolorjw.m: modified version of PCOLOR that expands the dimension the inputs X, Y and C so that the checkerboard will show all entries in C. The default PCOLOR clips the last row and column of C unless the shading('interp') is set, and the color of each square is determined by the value of C at the squares lower left corner. PCOLORJW recomputes X,Y at the mid points of the input, and pads C so that the effect is to shift each square one half space to the lower left. The perimeter squares are only half the width/height of all the others as can be seen if one sets shading('faceted'). Many thanks to John Wilkin for providing this modified function.
- matlab/utility/plot_nesting.m: plots requested ROMS nesting variable from input history NetCDF files. This function is very useful when debugging a ROMS nesting application. The plotting is not that fancy but it provides enough information for browsing ROMS variables very quickly.
- matlab/utility/plot_perimeter.m: adds a grid perimeter outline to an existing figure plotted with plot_nesting.m. It is used for browsing quickly output ROMS variables during nesting.
The above plot was done as follows:
Hnames={'scs_his_0010.nc', 'luzon_his_0010.nc'};
G=grids_structure(Hnames);
F=plot_nesting(G, Hnames, 'zeta', 8, 40, 1, [-2 4]);
shading flat; colormap(zebra);
h=plot_perimeter(G, 'r-');
print -r300 -dpng scs_ssh_day10.png
|
| Description |
nesting.F lines 850-855 has a call using DU_avg1 but it should DV_avg1:
CALL get_contact2d (dg, model, tile, &
& v2dvar, 'DV_avg1', &
& cr, Vcontact(cr)%Npoints, Vcontact, &
& LBi, UBi, LBj, UBj, &
& COUPLING(dg) % DV_avg1, &
& COMPOSITE(cr) % DV_avg1)
|

