| Description |
- Added the time-averaged switches (Aout) for biological and sediment quadratic tracers in their respective input scripts:
In bio_fennel.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields
! into AVERAGE output file: [1:NBT,Ngrids].
Aout(idTvar) == 12*T ! ..., NO3, ... biological tracer
Aout(idTTav) == 12*F ! ..., NO3_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == 12*F ! ..., u_NO3, ... quadratic <u*t> tracer terms
Aout(idVTav) == 12*F ! ..., v_NO3, ... quadratic <v*t> tracer terms
Aout(iHUTav) == 12*F ! ..., Huon_NO3, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == 12*F ! ..., Hvom_NO3, ... tracer volume flux, <Hvom*t>
In ecosim.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields
! into AVERAGE output file, [NBT,Ngrids] values are expected.
Aout(idTvar) == 61*T ! ..., DIC, ... biological tracer
Aout(idTTav) == 61*F ! ..., DIC_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == 61*F ! ..., u_DIC, ... quadratic <u*t> tracer terms
Aout(idVTav) == 61*F ! ..., v_DIC, ... quadratic <v*t> tracer terms
Aout(iHUTav) == 61*F ! ..., Huon_DIC, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == 61*F ! ..., Hvom_DIC, ... tracer volume flux, <Hvom*t>
In nemuro.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields
! into AVERAGE output file: [1;NBT,Ngrids].
Aout(idTvar) == 11*T ! ..., NO3, ... biological tracer
Aout(idTTav) == 11*F ! ..., NO3_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == 11*F ! ..., u_NO3, ... quadratic <u*t> tracer terms
Aout(idVTav) == 11*F ! ..., v_NO3, ... quadratic <v*t> tracer terms
Aout(iHUTav) == 11*F ! ..., Huon_NO3, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == 11*F ! ..., Hvom_NO3, ... tracer volume flux, <Hvom*t>
In npzd_Franks.in and npzd_Powell.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields
! into AVERAGE output file: [1:NBT,Ngrids].
Aout(idTvar) == 4*T ! ..., NO3, ... biological tracer
Aout(idTTav) == 4*F ! ..., NO3_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == 4*F ! ..., u_NO3, ... quadratic <u*t> tracer terms
Aout(idVTav) == 4*F ! ..., v_NO3, ... quadratic <v*t> tracer terms
Aout(iHUTav) == 4*F ! ..., Huon_NO3, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == 4*F ! ..., Hvom_NO3, ... tracer volume flux, <Hvom*t>
In npzd_iron.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged fields
! into AVERAGE output file: [1:NBT,Ngrids].
Aout(idTvar) == 6*T ! ..., NO3, ... biological tracer
Aout(idTTav) == 6*F ! ..., NO3_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == 6*F ! ..., u_NO3, ... quadratic <u*t> tracer terms
Aout(idVTav) == 6*F ! ..., v_NO3, ... quadratic <v*t> tracer terms
Aout(iHUTav) == 6*F ! ..., Huon_NO3, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == 6*F ! ..., Hvom_NO3, ... tracer volume flux, <Hvom*t>
In sediment.in we now have:
! Logical switches (TRUE/FALSE) to activate writing of time-averaged
! cohesive sediment fields into AVERAGE output file.
Aout(idmud) == T ! mud_01, ... suspended concentration
Aout(iMTTav) == F ! mud_01_2, ... quadratic <t*t> tracer terms
Aout(iMUTav) == F ! u_mud_01, ... quadratic <u*t> tracer terms
Aout(iMVTav) == F ! v_mud_01, ... quadratic <v*t> tracer terms
Aout(MHUTav) == F ! Huon_mud_01, ... tracer volume flux, <Huon*t>
Aout(MHVTav) == F ! Hvom_mud_01, ... tracer volume flux, <Hvom*t>
Aout(iMUbld) == T ! bedload_Umud_01, ... bed load at U-points
Aout(iMVbld) == T ! bedload_Vmud_01, ... bed load at V-points
...
! Logical switches (TRUE/FALSE) to activate writing of time-averaged
! non-cohesive sediment fields into AVERAGE output file.
Aout(idsand) == T ! sand_01, ... suspended concentration
Aout(iSTTav) == F ! sand_01_2, ... quadratic <t*t> tracer terms
Aout(iSUTav) == F ! u_sand_01, ... quadratic <u*t> tracer terms
Aout(iSVTav) == F ! v_sand_01, ... quadratic <v*t> tracer terms
Aout(SHUTav) == F ! Huon_sand_01, ... tracer volume flux, <Huon*t>
Aout(SHVTav) == F ! Hvom_sand_01, ... tracer volume flux, <Hvom*t>
Aout(iSUbld) == T ! bedload_Usand_01, ... bed load at U-points
Aout(iSVbld) == T ! bedload_Vsand_01, ... bed load at V-points
WARNING: The name of all tracers quadratic variables were renamed for clarity. And underscore was added to the basic variable. For example, the quadratic values for temperature are:
Aout(idTTav) == F F ! temp_2, ... quadratic <t*t> tracer terms
Aout(idUTav) == F F ! u_temp, ... quadratic <u*t> tracer terms
Aout(idVTav) == F F ! v_temp, ... quadratic <v*t> tracer terms
Aout(iHUTav) == F F ! Huon_temp, ... tracer volume flux, <Huon*t>
Aout(iHVTav) == F F ! Hvom_temp, ... tracer volume flux, <Hvom*t>
Examples of the variable names for biology and sediment are shown above.
Many thanks to Aboozar Tabatabai for bringing this to my attention.
WARNING: Notice that all the input scripts for physics (ocean.in) in addition to the ones for biology and sediment were updated. The changes to the ocean.in are only done in the documentation for Aout. The test cases repository is also updated accordingly.
|
| Description |
This is an important update for the nesting algorithms:
- Corrected bugs in subroutine put_refine2d of nesting.F. The conditional to avoid updating contact points at refined grid physical boundaries for time index indx1(ng) was incorrect.
- Corrected the time index to t2 for the donor grid mass fluxes REFINED(cr)%DU_avg2(1,m,t2) and REFINED(cr)%DV_avg2(1,m,t2) in put_refine2d.
- Corrected the logic to use more than one refined grid in an application. The logic to initialize arrays in structure BRY_CONTACT(:,cr) is more robust by introducing the conjugate contact region (CCR) concept. See mod_nesting.F for details.
- Corrected the index when the REFINED structure is used. Changed REFINED(ir) to REFINED(cr). This was a bug when we have more than one refined grid in an application.
- Corrected the reading of several variables for the nested grid connectivity NetCDF file, NGCNAME, in set_contact.F. The local variables Lcoincident, Lcomposite, Lmosaic, and Lrefinement need to be dimension (Ngrids-1)*2 and not Ngrids. That is, they need to be of the size of number of contact regions, Ncontact = (Ngrids-1)*2. This was also corrected in the Matlab scripts c_contact.m and wrt_contact.m that defines and writes such variables.
WARNING: Users need to also update the Matlab repository and recreate the nesting grid connectivity NetCDF file, NGCNAME, for their applications.
Also correct few things in other files:
- Added a more robust logic in the vertical interpolation in the 4D-Var routines extract_obs.F and ad_extract_obs.F. Many thanks to Paul Mattern for bringing this to my attention.
Comments on the nesting algorithms:
- We are still learning how to use these algorithms and designing simple applications to insure that algorithms were coded correctly for the intended capabilities. We found out that in order to conserve volume in refinement applications, the nesting has to be two-way. This is a very interesting result that we are trying to find complete explanations for it. Our applications with one-way nesting (ONE_WAY) tend to loose volume after several days.
This can be illustrated by adding two refined grids to the LAKE_SIGNELL application:
One-way, free-surface after 2 days of simulation:
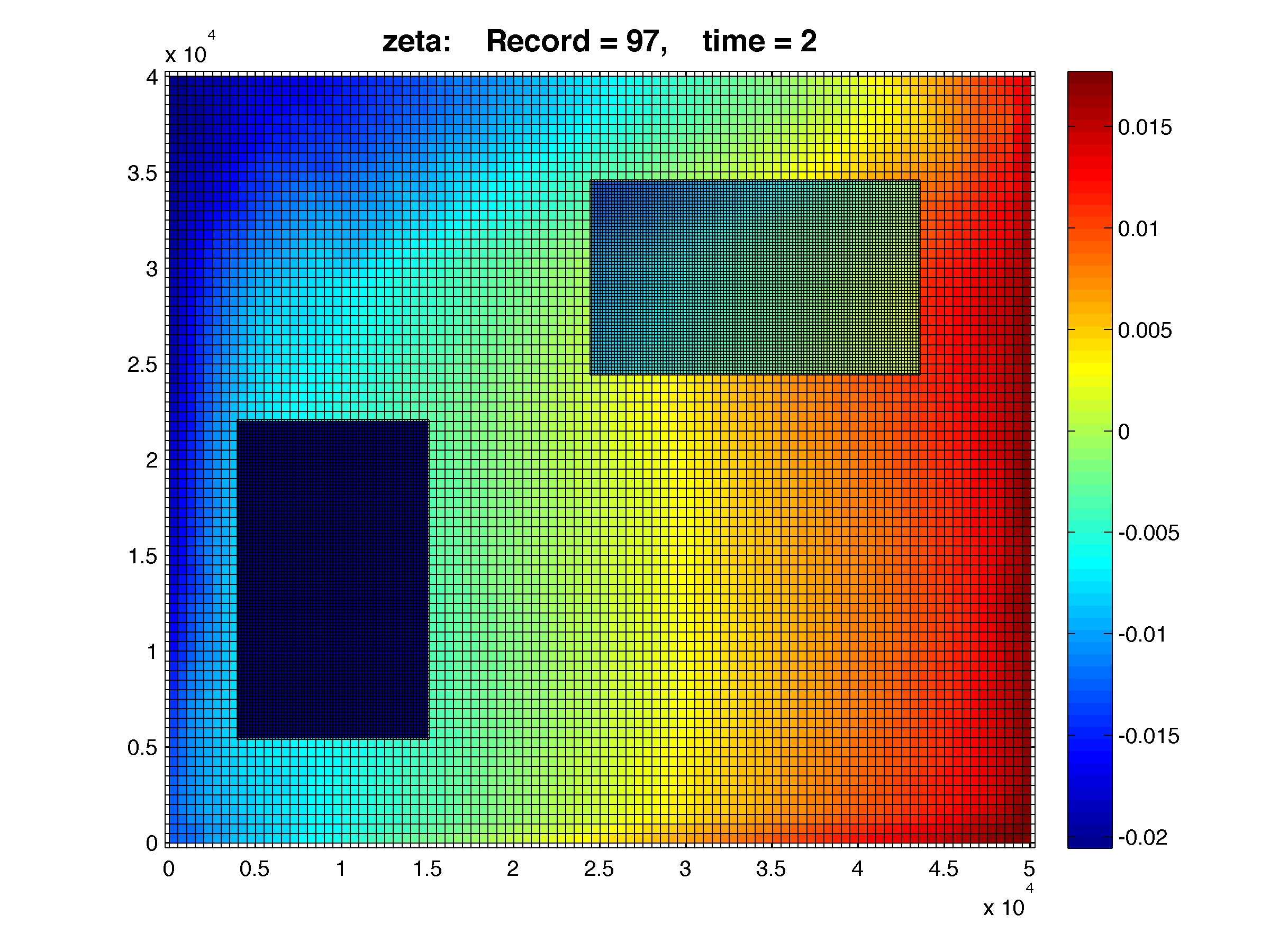
Notice that the free-surface in the bottom-left and top-right refined grids have lower values when compared to the coarse grid values. The refinement is 1:3 for both finer grids. The bathymetry is flat. Here both refined grids are loosing volume slowly in time. The bottom-left grid is completely dark blue. The top-right grid exhibits much lower values. Just click on the figure to see a large resolution figure.
Two-way, free-surface after 2 days of simulation:
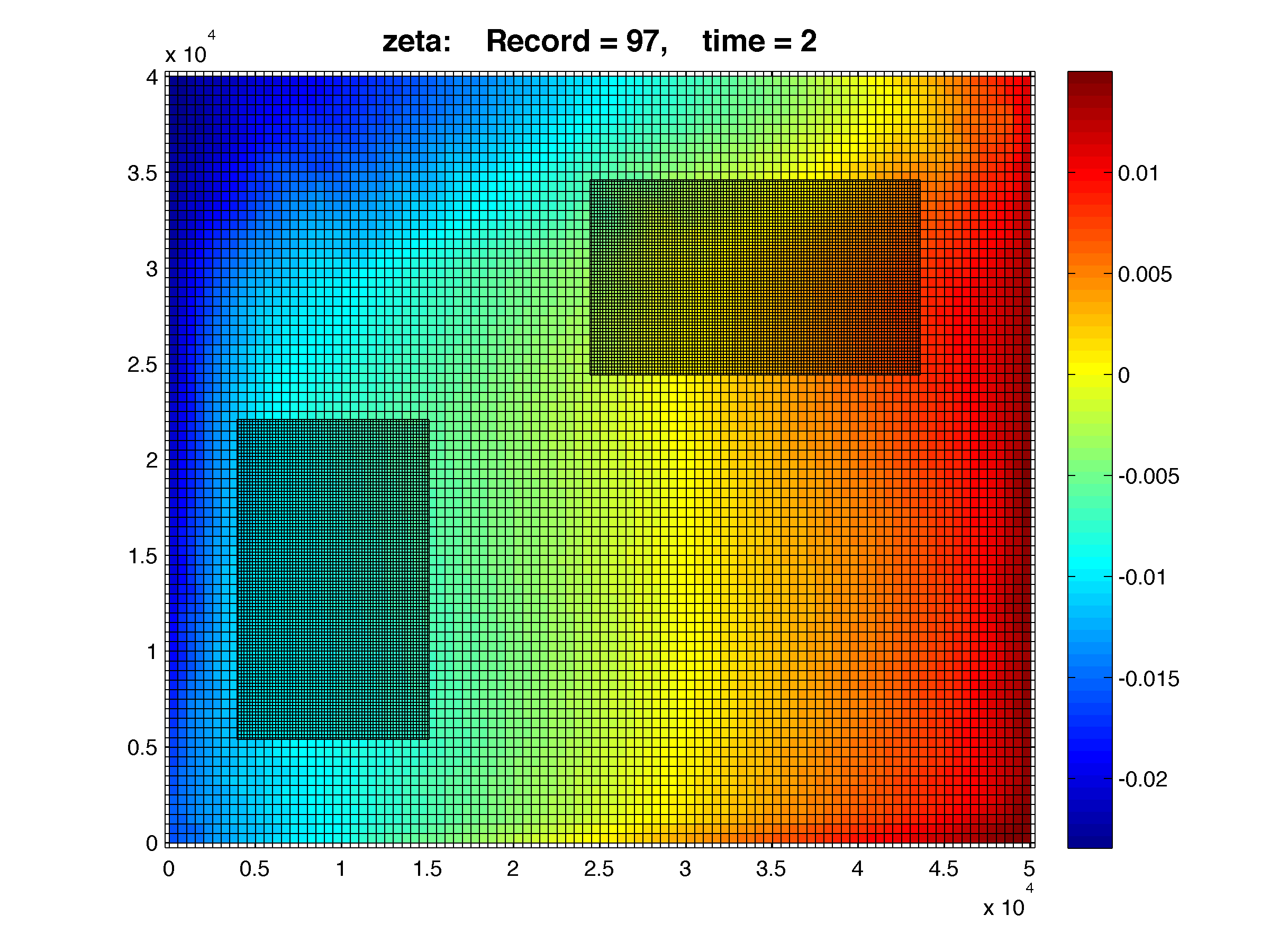
This is the same configuration as the above case but with ONE_WAY turned off. That is, ROMS is run in a two-way configuration. You can see the smooth continuation of colors between the coarse and finer grids. Notice the finer grids have additional physics since the gravity wave phenomena is better resolved. In our realistic applications we can see the trail of internal waves.
I will load the LAKE_SIGNELL nesting application to the test repository soon. Many thanks to John Wilkin for setting this grids and testing the nesting algorithms. We are now looking at the science of nesting instead of coding...
- It is very clear from our experience, so far, that we need to pay a lot of attention to how the nested grids and the connectivity information is built. We need to conserve volume and compute the bathymetry accordingly... I foresee the Matlab scripts to continue to evolve as we gain more experience and try new applications.
|


