Custom Query (964 matches)
Results (826 - 828 of 964)
| Ticket | Owner | Reporter | Resolution | Summary |
|---|---|---|---|---|
| #812 | Fixed | Updated River Runoff: Option LuvSrc | ||
| Description |
In the ROMS Forum, Julia Levin proposed some enhancements to the Point Source algorithm to model river runoff using either the standard input switches LuvSrc or LwSrc. The LuvSrc option is used to model river runoff transport by modifying the horizontal advection transport fluxes whereas the LwSrc option is used as a vertical mass volume inflow. I updated inp_decode.F to be more robust with the gfortran compiler during debugging. Really weird behavior in the DO-loop counter. It continued looping after the specified range. No idea where that comes from but this update force a strict looping. For example, in load_1d_i, we now have: ! If not all values are provided for the variable, assume the last value
! for the rest of the array.
!
ic=0
IF (Ninp.le.Nout) THEN
DO i=1,Ninp
ic=ic+1
Vout(i)=INT(Vinp(i))
END DO
IF (Nout.gt.Ninp) THEN
Nstr=Ninp+1
DO i=Nstr,Nout
ic=ic+1
Vout(i)=INT(Vinp(Ninp))
END DO
END IF
ELSE
DO i=1,Nout
ic=ic+1
Vout(i)=INT(Vinp(i))
END DO
END IF
Nval=ic
I only implemented the changes associated with LuvSrc. The suggested changes for LwSrc are unstable and blow-up for the TS_MPDATA advection option. It requires further work. The old algorithm implemented the vertical mass inflow by modifying the vertical advection fluxes in the tracer equations and by modifying the omega equation that is associated with the 3D continuity equation. The proposed change is to introduce a forcing term to the rate of change of tracer. At the moment, such treatment is problematic and unstable for a simple Euler forward step. There are other issues to consider like conservation and variance of the tracer. For the LuvSrc, we now have in pre_step3d.F: !
! Apply tracers point sources to the horizontal advection terms,
! if any.
!
IF (LuvSrc(ng)) THEN
DO is=1,Nsrc(ng)
Isrc=SOURCES(ng)%Isrc(is)
Jsrc=SOURCES(ng)%Jsrc(is)
IF (((Istr.le.Isrc).and.(Isrc.le.Iend+1)).and. &
& ((Jstr.le.Jsrc).and.(Jsrc.le.Jend+1))) THEN
IF (INT(SOURCES(ng)%Dsrc(is)).eq.0) THEN
IF (LtracerSrc(itrc,ng)) THEN
FX(Isrc,Jsrc)=Huon(Isrc,Jsrc,k)* &
& SOURCES(ng)%Tsrc(is,k,itrc)
ELSE
FX(Isrc,Jsrc)=0.0_r8
END IF
ELSE
IF (LtracerSrc(itrc,ng)) THEN
FE(Isrc,Jsrc)=Hvom(Isrc,Jsrc,k)* &
& SOURCES(ng)%Tsrc(is,k,itrc)
ELSE
FE(Isrc,Jsrc)=0.0_r8
END IF
END IF
END IF
END DO
END IF
Changes were made to the tangent, representer, and adjoint versions of pre_step3d.F and step3d_t.F. Many thanks to Julia Levin, John Wilkin, and Chuning Wang (Jupiter Intelligence INC) for looking and testing the formulation of Point Sources. |
|||
| #814 | Done | IMPORTANT: EcoSim Bio-Optical Diagnostic Terms | ||
| Description |
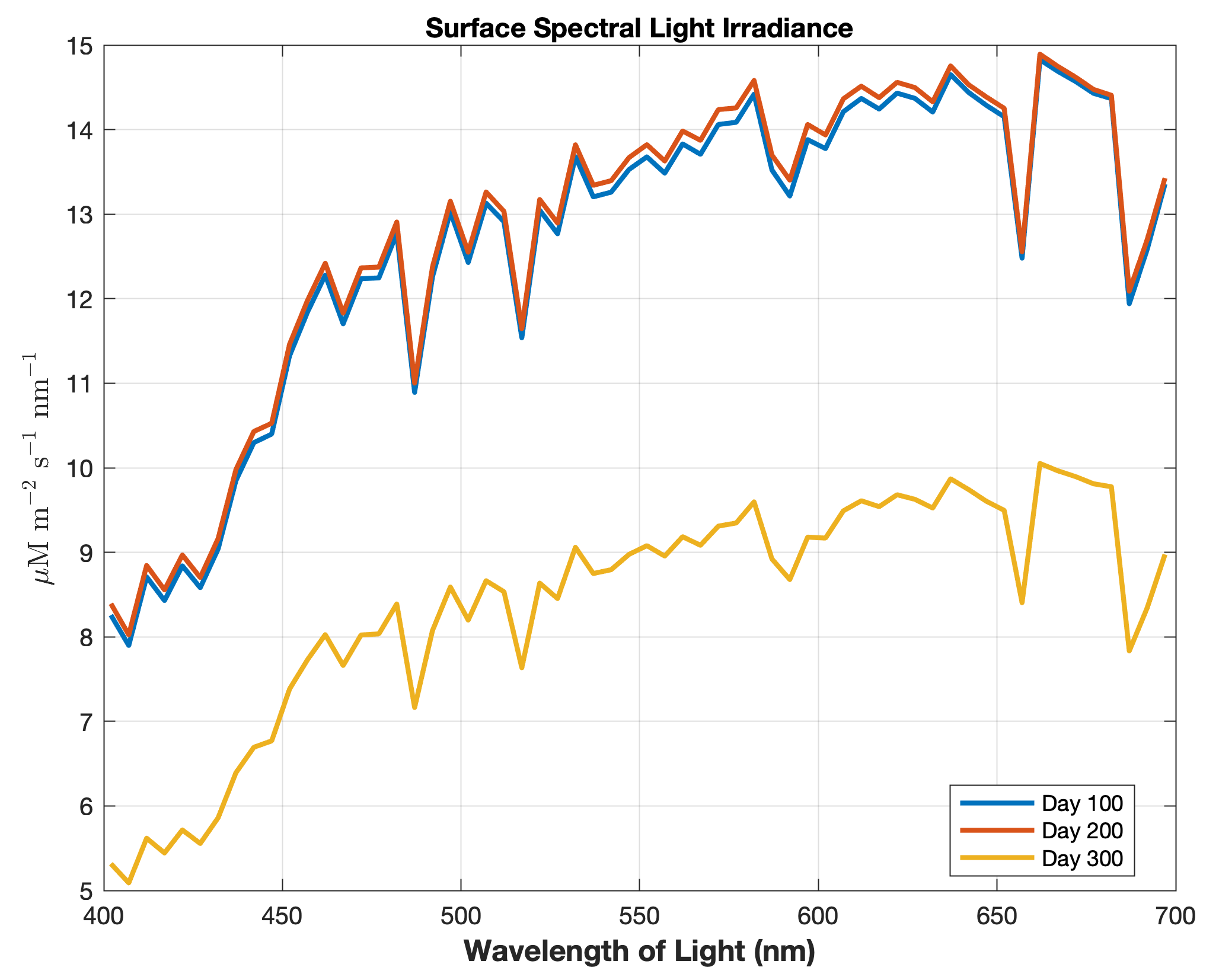
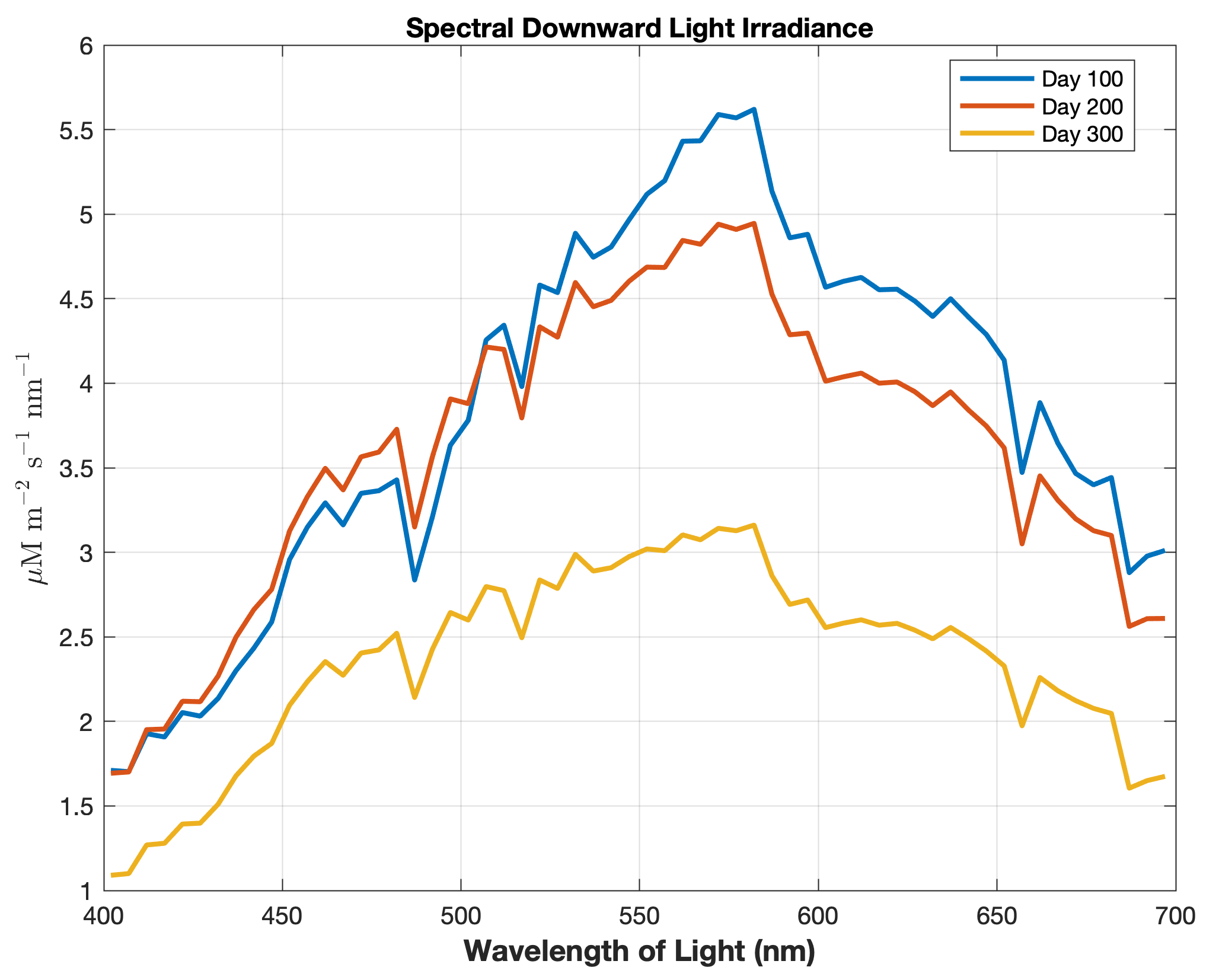
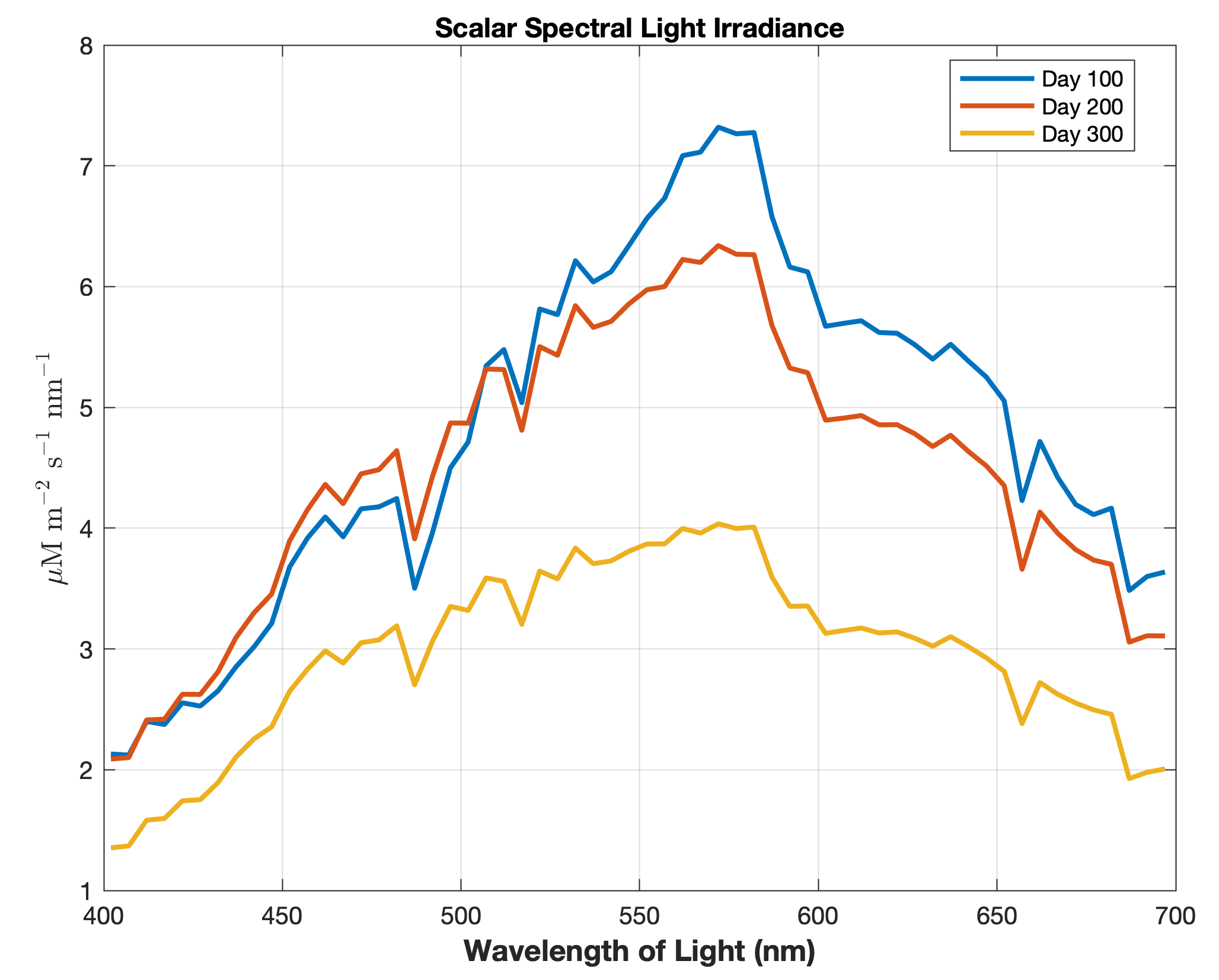
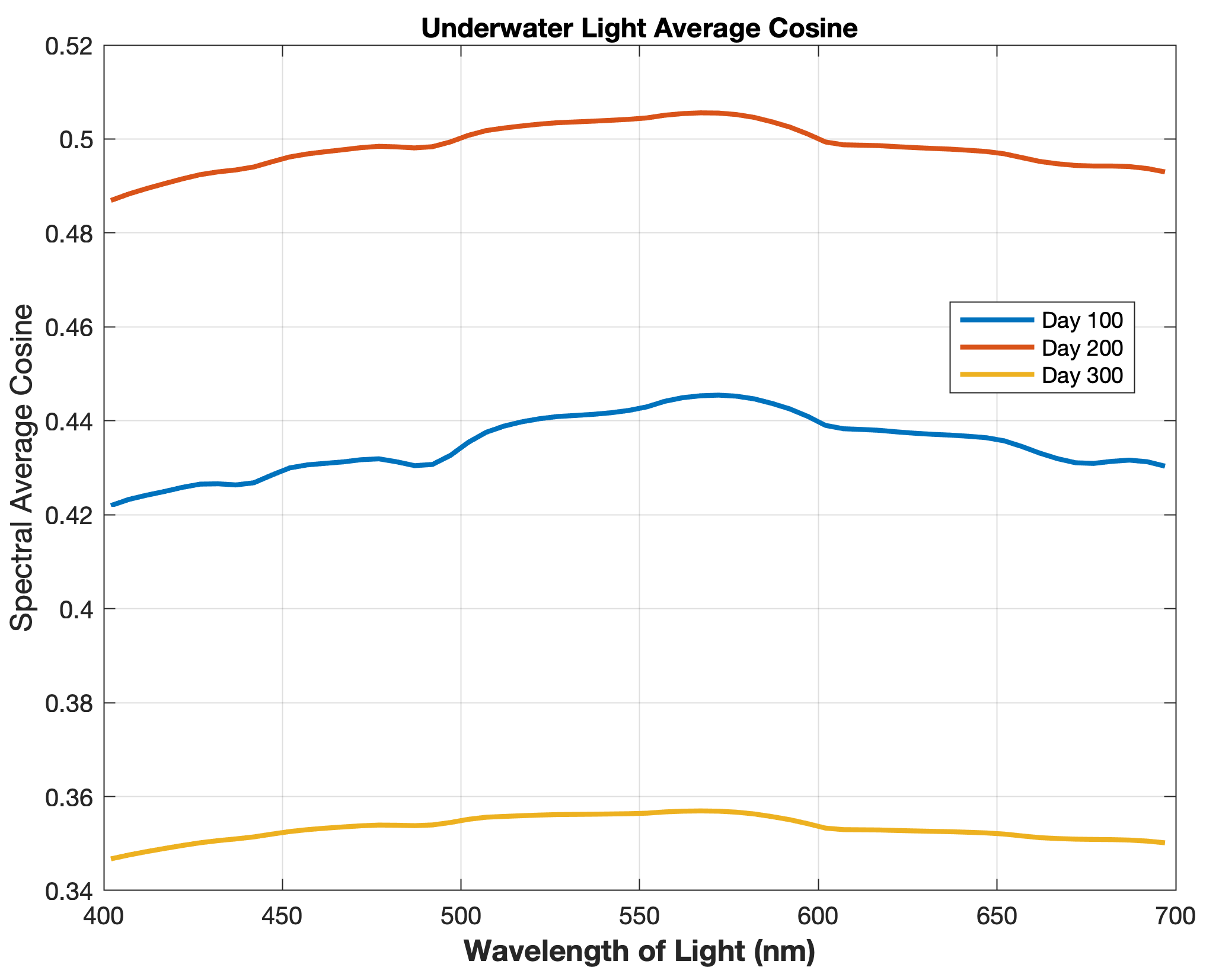
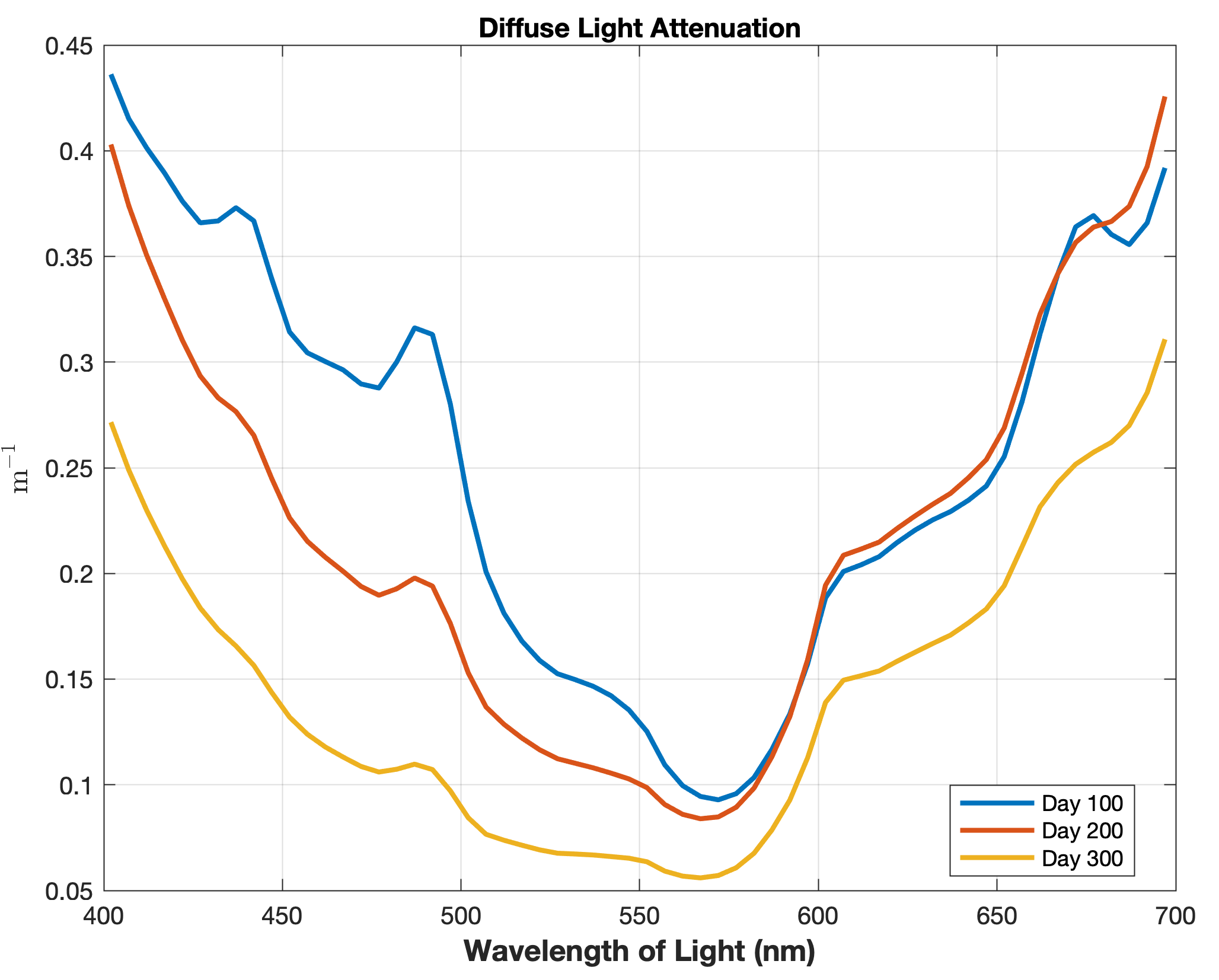
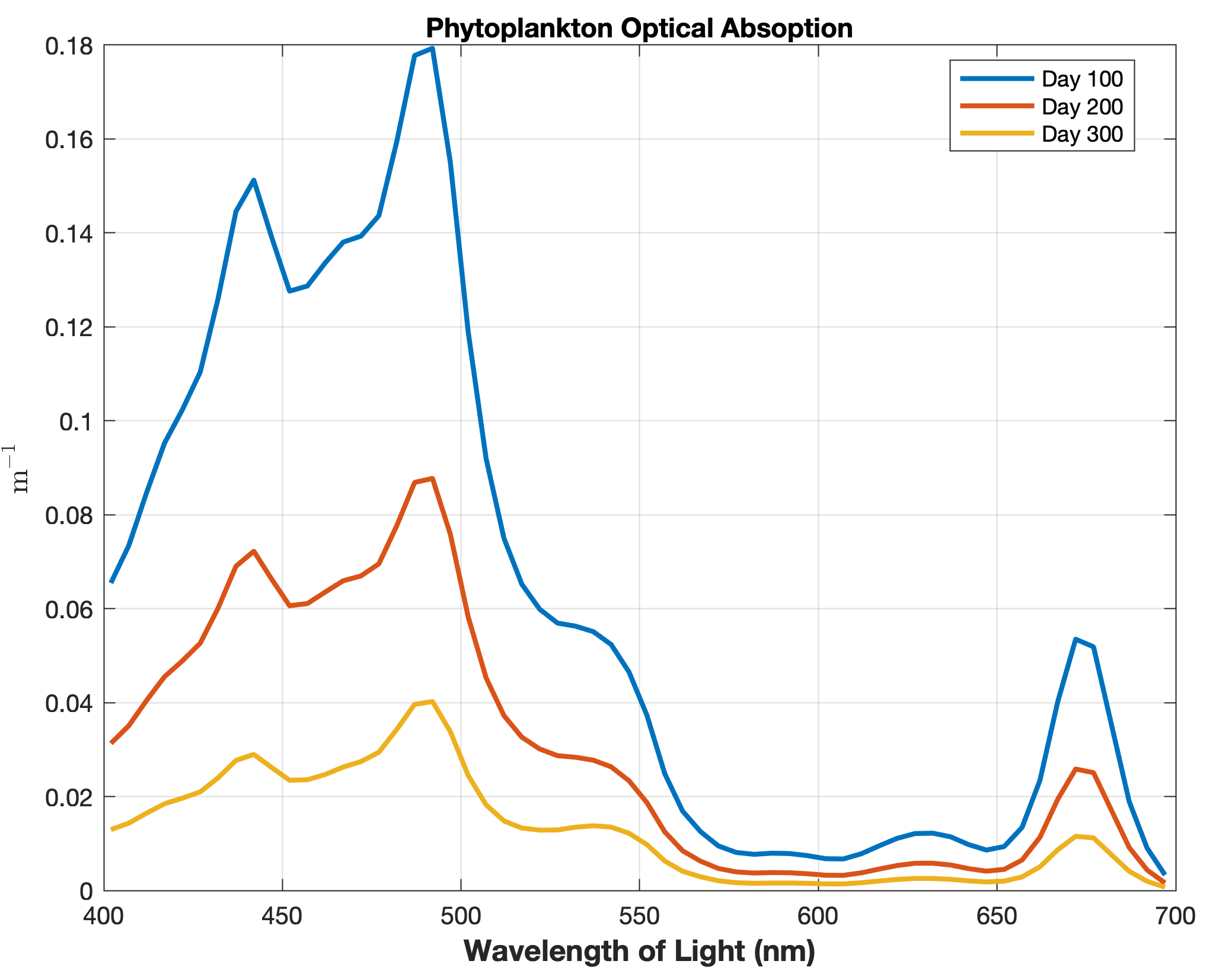
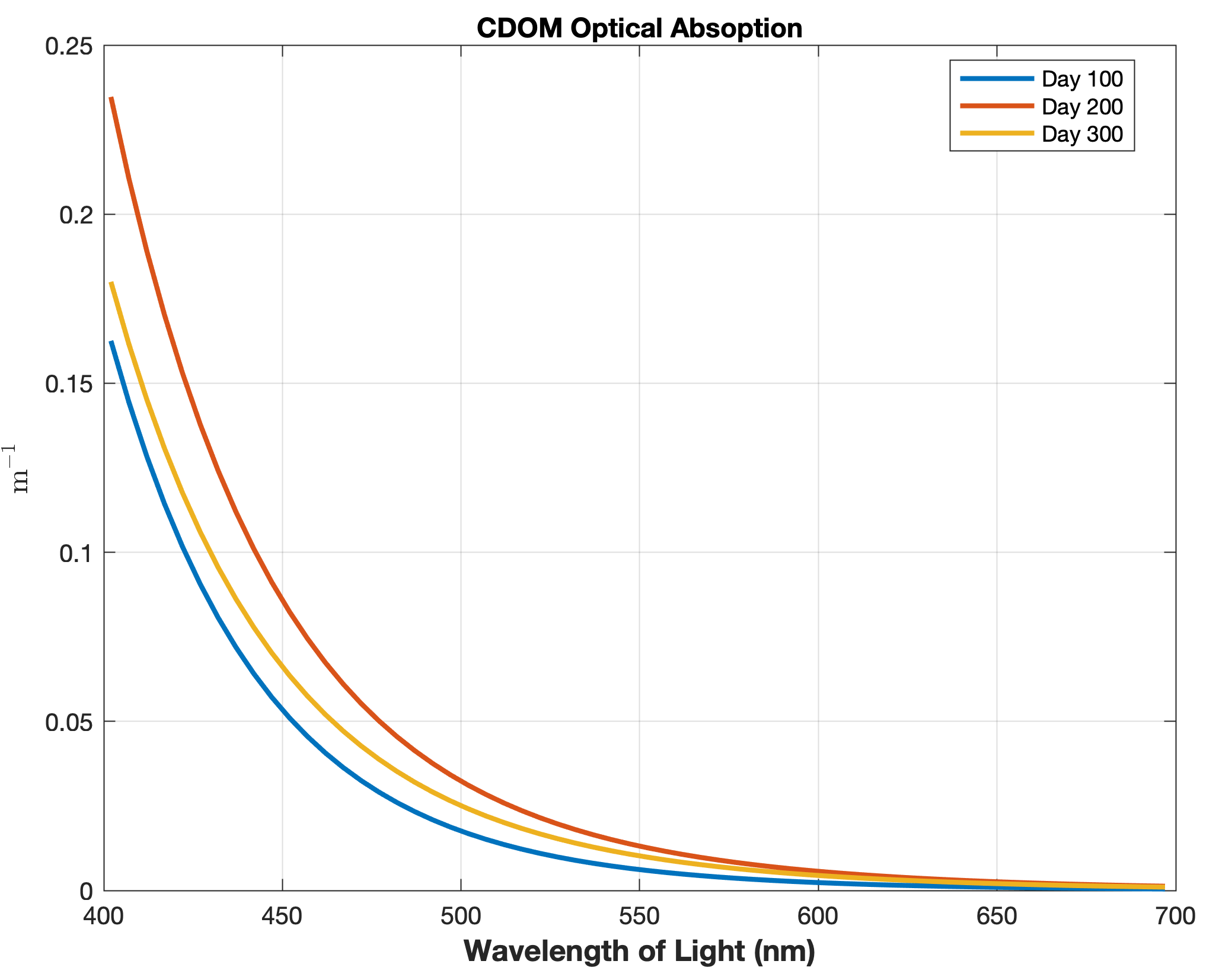
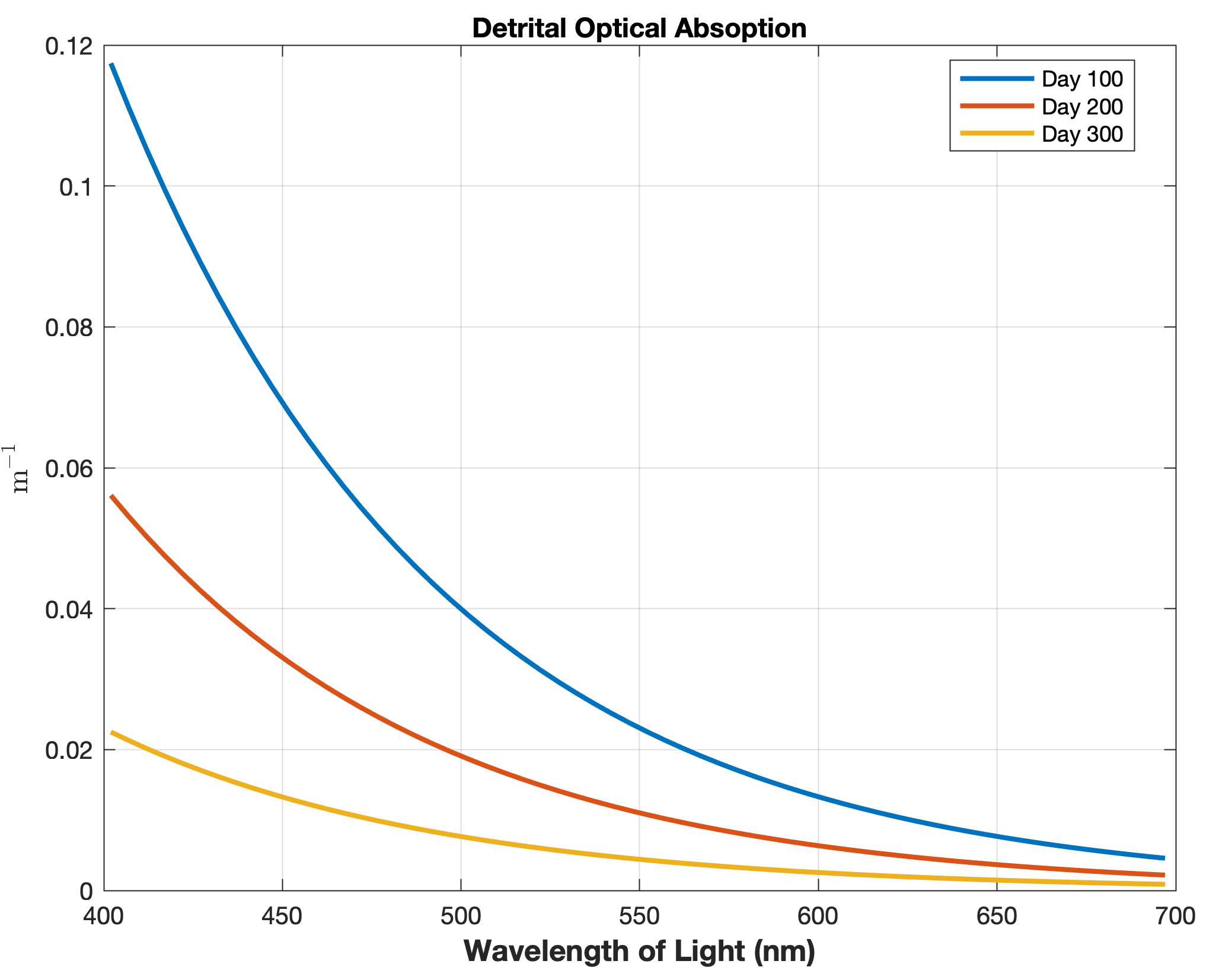
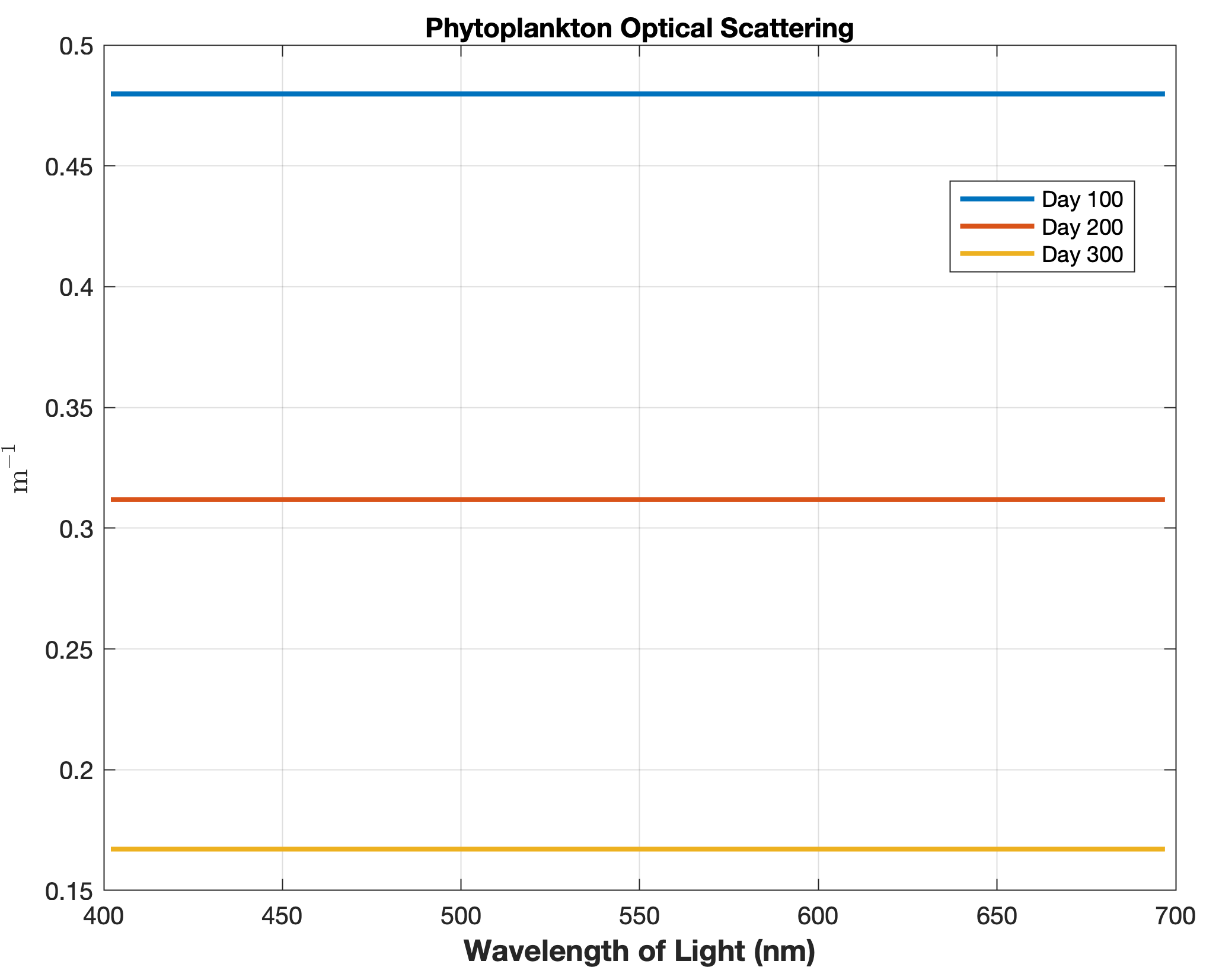
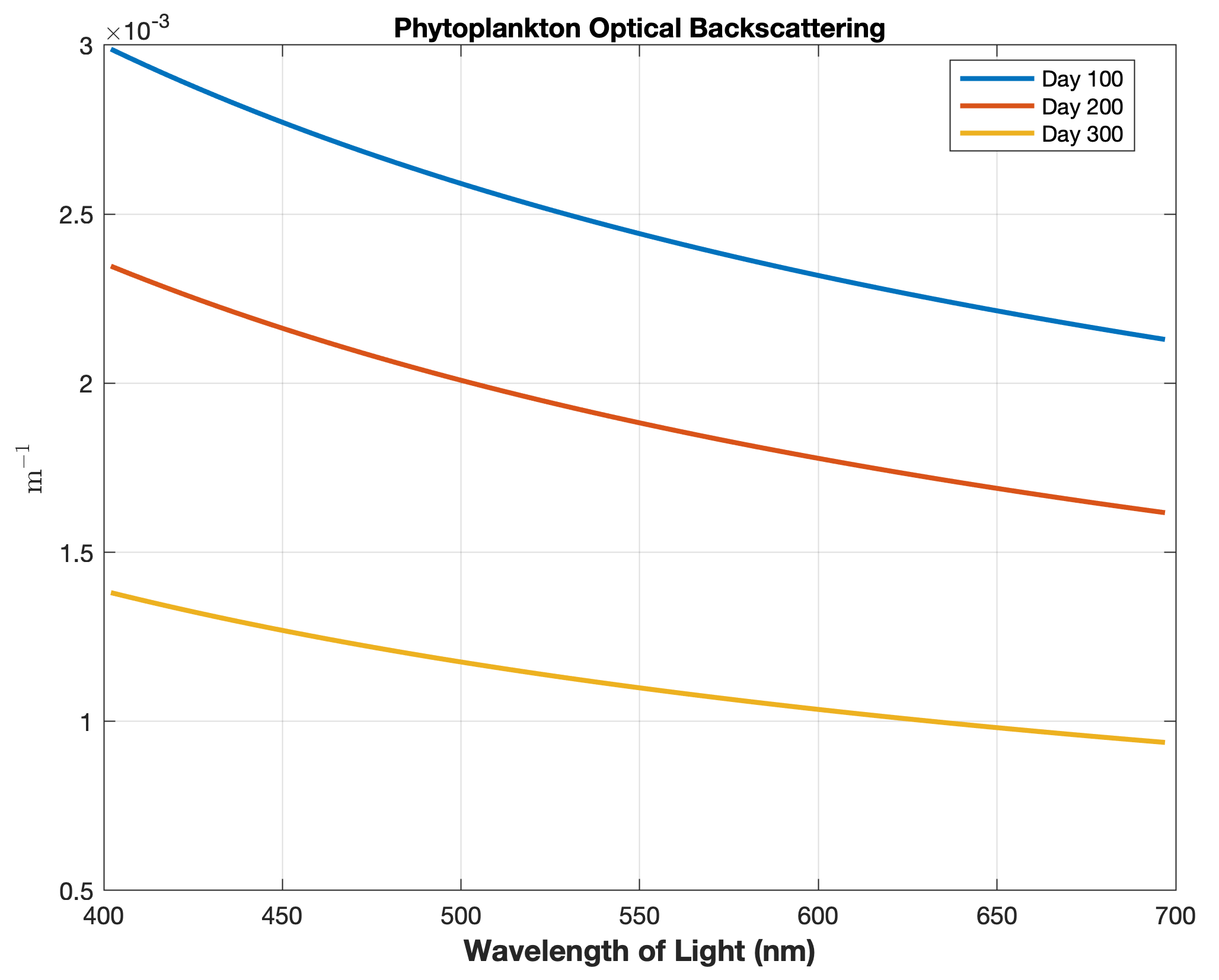
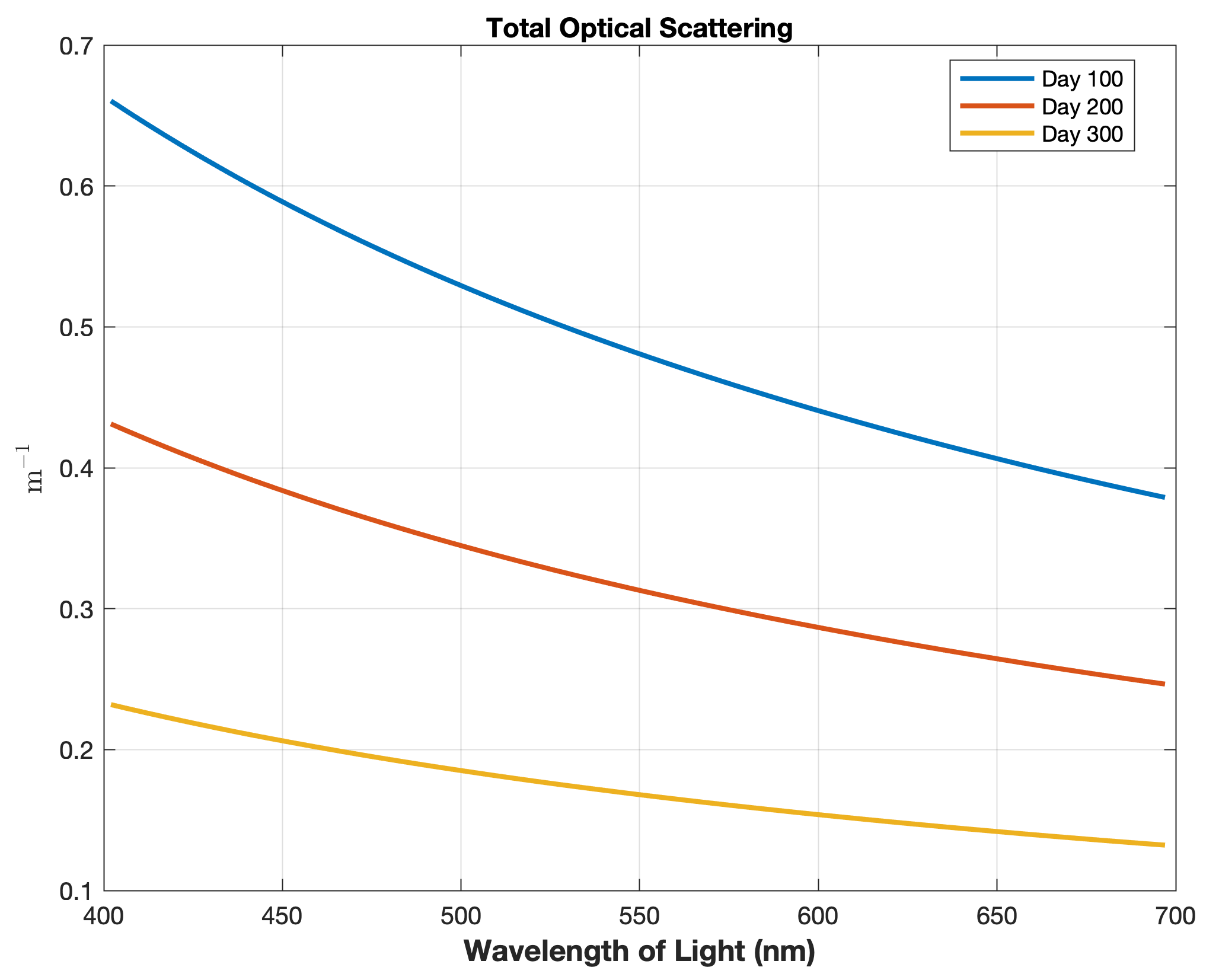
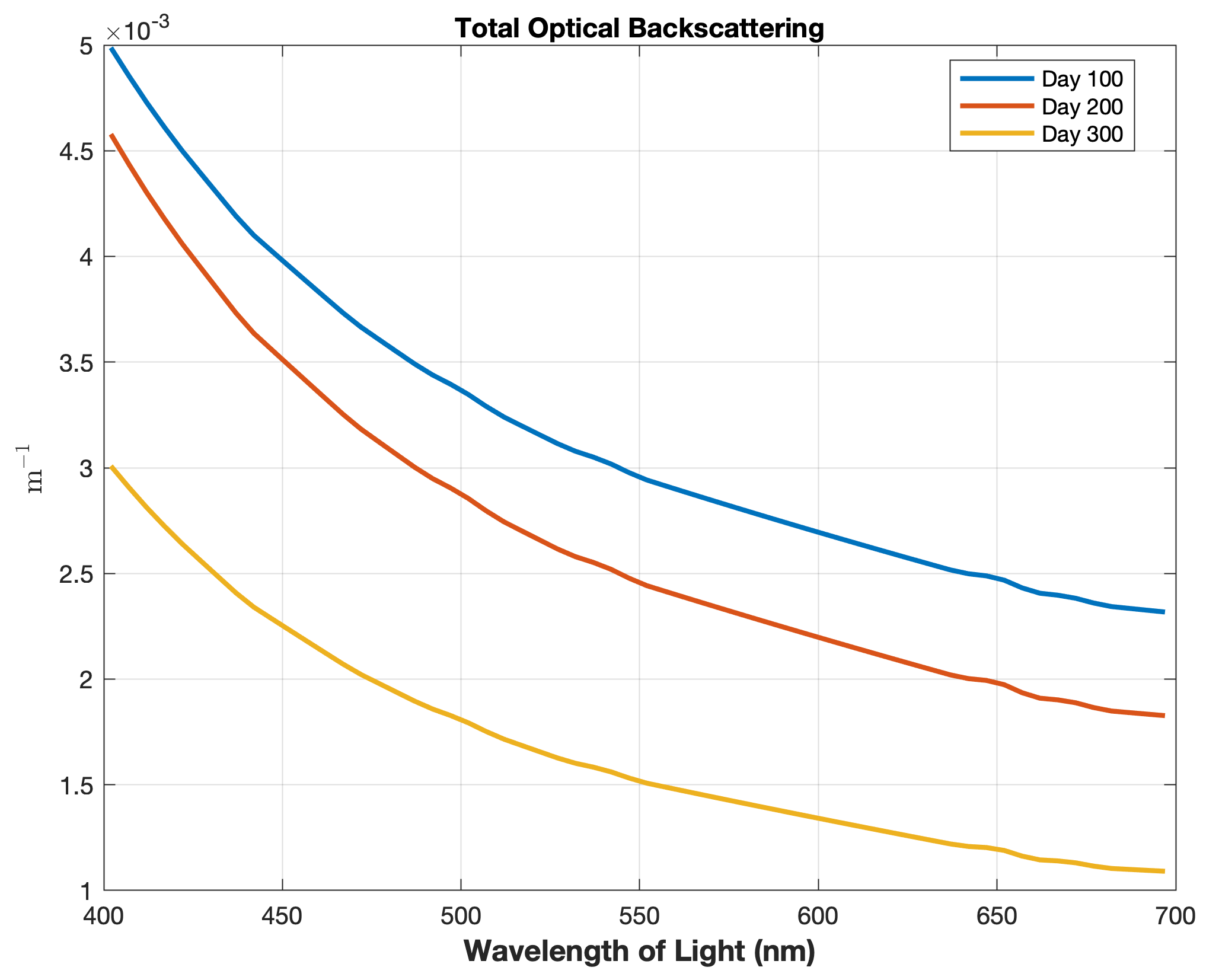
Updated the EcoSim model to include diagnostics of the optical terms when DIAGNOSTICS_BIO and BIO_OPTIC to write out into output NetCDF file DIANAME various underwater light spectral properties.
Many thanks to Bronwyn Cahill for her help coding and testing this update to the EcoSim model. |
|||
| #815 | Done | ROMS GIT and SVN Repositories for version control | ||
| Description |
ROMS now supports both GIT and SVN version control repositories to check out and update the source code. Both GIT and SVN have their own trac wiki to manage the code and browse updates and bug tracking. However, update or bug trac tickets need to be posted in the SVN trac wiki, for now.
Both repositories use the same username and password selected during ROMS registration. It is a matter of preference what repository system the user selects to managing the code, and we are not recommending which one to use. The ROMS code is identical in both repositories. For detailed information check WikiROMS pages for GIT and SVN usage: The code will be updated later to differentiate specific information between GIT and SVN version identifiers like IDs, NetCDF files and standard output metadata, and the Version file. |
|||