1. I was asked to help Ken Coyle with a problem in the GOANPZ code. It turned out to be a missing chunk in inp_par.F, converting the boundary nudging timescale from units of days to sec-1. This code is duplicated for each biology model and was missing for the GOA version – could this be cleaned up so that such duplication isn’t needed?
2. I was getting odd differences in the ice depending on my domain tiling. I tracked it down to code in the ICE part of bulk_flux.F, setting the surface albedo if there is ice, leaving it random otherwise. I also took out the fice variable since it was just used the once (and not being set right either).
3. Of more general interest, I tried out the new code for interpolating from coarse forcing files. I was surprised to get stress fields such as:
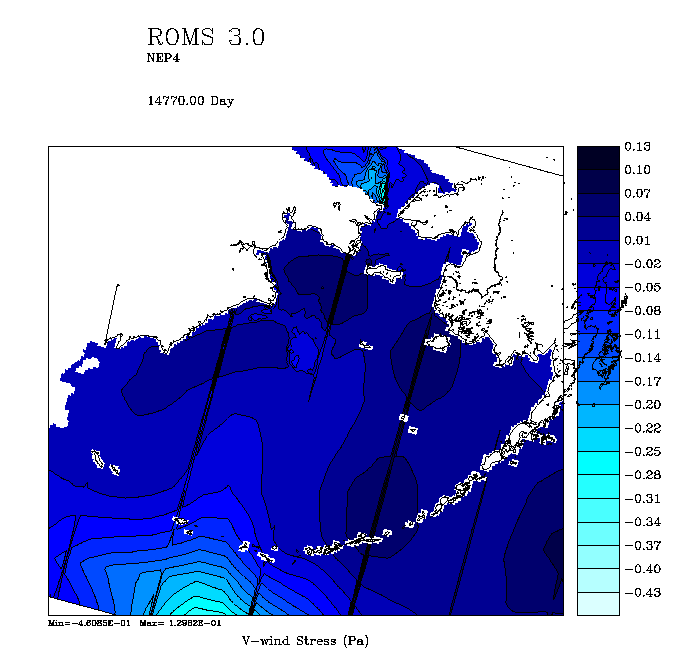
The operation order is that the coarse grid data is read in and interpolated to the ROMS grid during get_data.F. It is then time-interpolated, exchanged, and finally rotated in set_data.F. It seems to me that it would be more efficient to rotate it in get_data.F since that is only happening every time you read the (six hourly) winds, not every timestep. I’m off to try to fix this.
Just a nit on the CPP option around the rotation code: CURVGRID is for when the dmde and dndx terms are nonzero and require the extra advection terms to be computed. It is not a flag for whether your grid is aligned north-south and east-west. In fact, north-south grids generally have nonzero dmde on a sphere.
Edit: I think it’s working! Just a note that if you look at my get_data.F, it is just a bit different because Don Stark asked me to change it (and change who calls it) for the CCSM coupling effort at NCAR. Most important, he asked me to put the initialization stuff elsewhere (now in get_data_init.F).